Gilles Wainrib
ToxicBlend: Virtual Screening of Toxic Compounds with Ensemble Predictors
Jun 12, 2018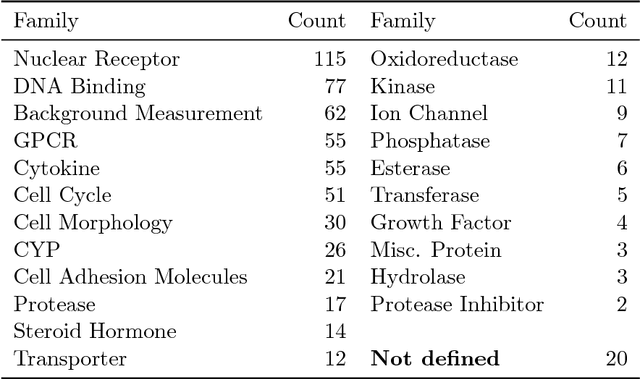
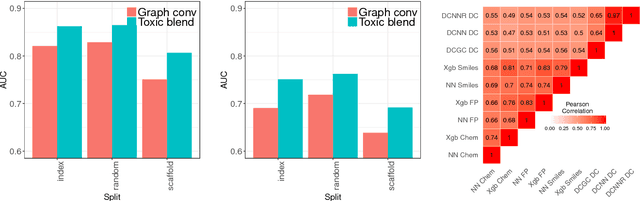
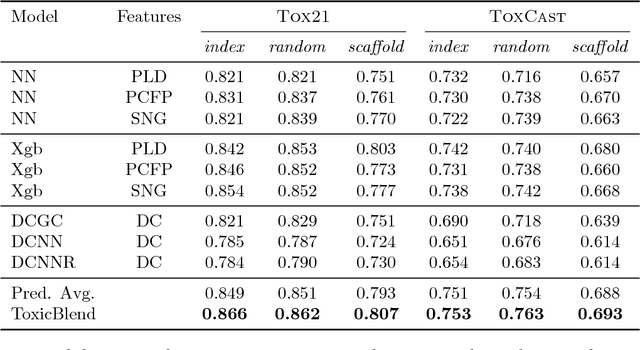
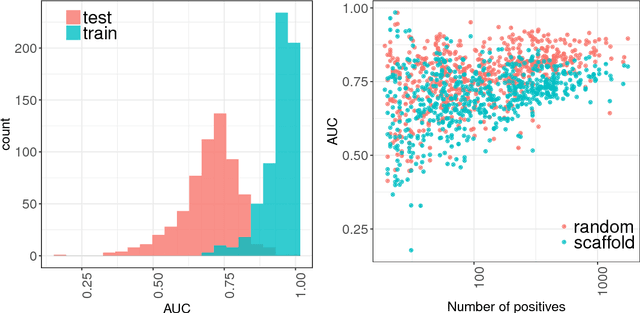
Abstract:Timely assessment of compound toxicity is one of the biggest challenges facing the pharmaceutical industry today. A significant proportion of compounds identified as potential leads are ultimately discarded due to the toxicity they induce. In this paper, we propose a novel machine learning approach for the prediction of molecular activity on ToxCast targets. We combine extreme gradient boosting with fully-connected and graph-convolutional neural network architectures trained on QSAR physical molecular property descriptors, PubChem molecular fingerprints, and SMILES sequences. Our ensemble predictor leverages the strengths of each individual technique, significantly outperforming existing state-of-the art models on the ToxCast and Tox21 toxicity-prediction datasets. We provide free access to molecule toxicity prediction using our model at http://www.owkin.com/toxicblend.
Scaling up Echo-State Networks with multiple light scattering
Feb 13, 2018



Abstract:Echo-State Networks and Reservoir Computing have been studied for more than a decade. They provide a simpler yet powerful alternative to Recurrent Neural Networks, every internal weight is fixed and only the last linear layer is trained. They involve many multiplications by dense random matrices. Very large networks are difficult to obtain, as the complexity scales quadratically both in time and memory. Here, we present a novel optical implementation of Echo-State Networks using light-scattering media and a Digital Micromirror Device. As a proof of concept, binary networks have been successfully trained to predict the chaotic Mackey-Glass time series. This new method is fast, power efficient and easily scalable to very large networks.
Classification and Disease Localization in Histopathology Using Only Global Labels: A Weakly-Supervised Approach
Feb 01, 2018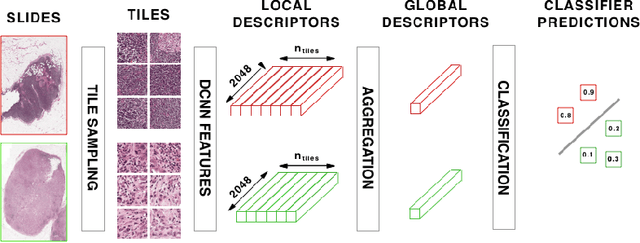
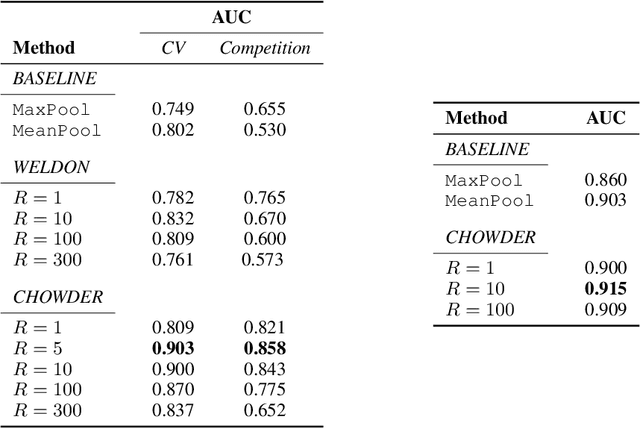
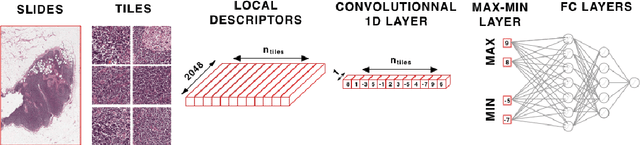
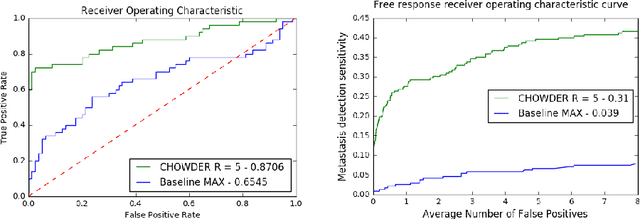
Abstract:Analysis of histopathology slides is a critical step for many diagnoses, and in particular in oncology where it defines the gold standard. In the case of digital histopathological analysis, highly trained pathologists must review vast whole-slide-images of extreme digital resolution ($100,000^2$ pixels) across multiple zoom levels in order to locate abnormal regions of cells, or in some cases single cells, out of millions. The application of deep learning to this problem is hampered not only by small sample sizes, as typical datasets contain only a few hundred samples, but also by the generation of ground-truth localized annotations for training interpretable classification and segmentation models. We propose a method for disease localization in the context of weakly supervised learning, where only image-level labels are available during training. Even without pixel-level annotations, we are able to demonstrate performance comparable with models trained with strong annotations on the Camelyon-16 lymph node metastases detection challenge. We accomplish this through the use of pre-trained deep convolutional networks, feature embedding, as well as learning via top instances and negative evidence, a multiple instance learning technique from the field of semantic segmentation and object detection.
Robust Detection of Covariate-Treatment Interactions in Clinical Trials
Dec 21, 2017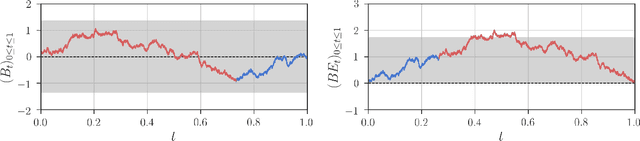

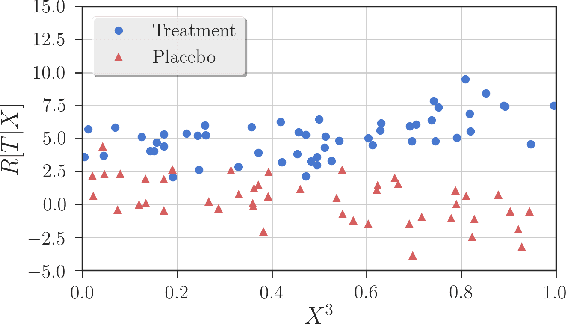
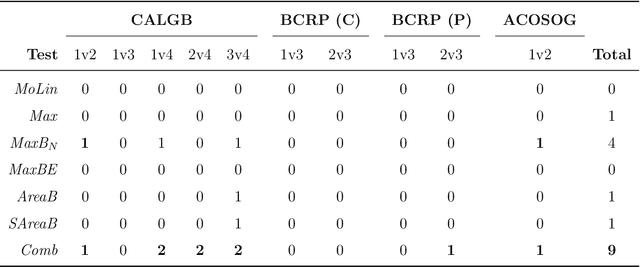
Abstract:Detection of interactions between treatment effects and patient descriptors in clinical trials is critical for optimizing the drug development process. The increasing volume of data accumulated in clinical trials provides a unique opportunity to discover new biomarkers and further the goal of personalized medicine, but it also requires innovative robust biomarker detection methods capable of detecting non-linear, and sometimes weak, signals. We propose a set of novel univariate statistical tests, based on the theory of random walks, which are able to capture non-linear and non-monotonic covariate-treatment interactions. We also propose a novel combined test, which leverages the power of all of our proposed univariate tests into a single general-case tool. We present results for both synthetic trials as well as real-world clinical trials, where we compare our method with state-of-the-art techniques and demonstrate the utility and robustness of our approach.
A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series
Nov 27, 2017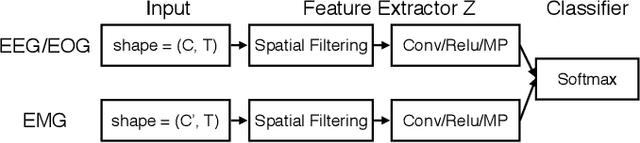
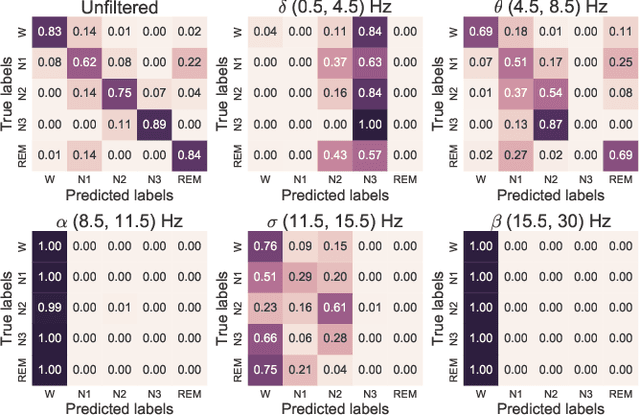
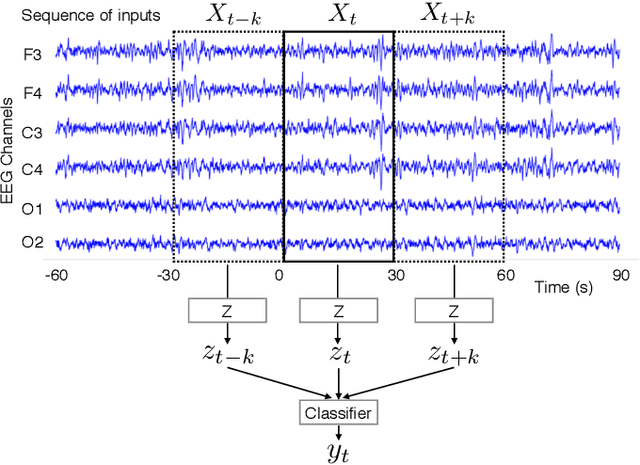
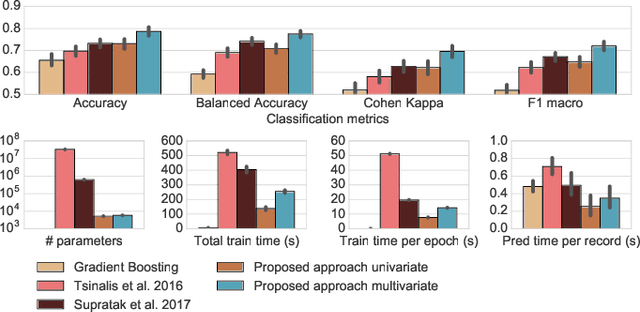
Abstract:Sleep stage classification constitutes an important preliminary exam in the diagnosis of sleep disorders. It is traditionally performed by a sleep expert who assigns to each 30s of signal a sleep stage, based on the visual inspection of signals such as electroencephalograms (EEG), electrooculograms (EOG), electrocardiograms (ECG) and electromyograms (EMG). We introduce here the first deep learning approach for sleep stage classification that learns end-to-end without computing spectrograms or extracting hand-crafted features, that exploits all multivariate and multimodal Polysomnography (PSG) signals (EEG, EMG and EOG), and that can exploit the temporal context of each 30s window of data. For each modality the first layer learns linear spatial filters that exploit the array of sensors to increase the signal-to-noise ratio, and the last layer feeds the learnt representation to a softmax classifier. Our model is compared to alternative automatic approaches based on convolutional networks or decisions trees. Results obtained on 61 publicly available PSG records with up to 20 EEG channels demonstrate that our network architecture yields state-of-the-art performance. Our study reveals a number of insights on the spatio-temporal distribution of the signal of interest: a good trade-off for optimal classification performance measured with balanced accuracy is to use 6 EEG with 2 EOG (left and right) and 3 EMG chin channels. Also exploiting one minute of data before and after each data segment offers the strongest improvement when a limited number of channels is available. As sleep experts, our system exploits the multivariate and multimodal nature of PSG signals in order to deliver state-of-the-art classification performance with a small computational cost.
The Asymptotic Performance of Linear Echo State Neural Networks
Mar 25, 2016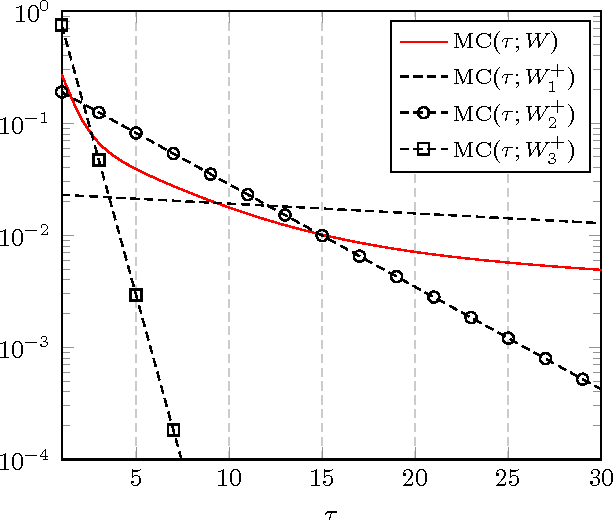
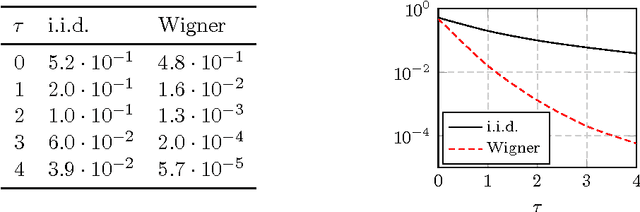
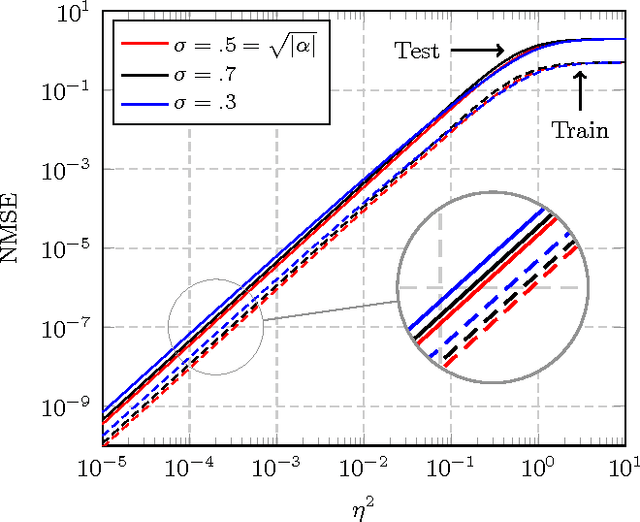
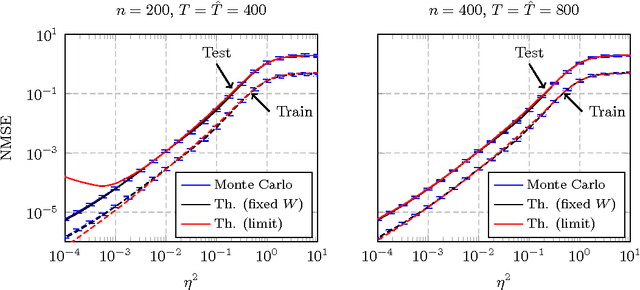
Abstract:In this article, a study of the mean-square error (MSE) performance of linear echo-state neural networks is performed, both for training and testing tasks. Considering the realistic setting of noise present at the network nodes, we derive deterministic equivalents for the aforementioned MSE in the limit where the number of input data $T$ and network size $n$ both grow large. Specializing then the network connectivity matrix to specific random settings, we further obtain simple formulas that provide new insights on the performance of such networks.
A biological gradient descent for prediction through a combination of STDP and homeostatic plasticity
Jun 11, 2013Abstract:Identifying, formalizing and combining biological mechanisms which implement known brain functions, such as prediction, is a main aspect of current research in theoretical neuroscience. In this letter, the mechanisms of Spike Timing Dependent Plasticity (STDP) and homeostatic plasticity, combined in an original mathematical formalism, are shown to shape recurrent neural networks into predictors. Following a rigorous mathematical treatment, we prove that they implement the online gradient descent of a distance between the network activity and its stimuli. The convergence to an equilibrium, where the network can spontaneously reproduce or predict its stimuli, does not suffer from bifurcation issues usually encountered in learning in recurrent neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge