Pierre Courtiol
Self-Supervision Closes the Gap Between Weak and Strong Supervision in Histology
Dec 07, 2020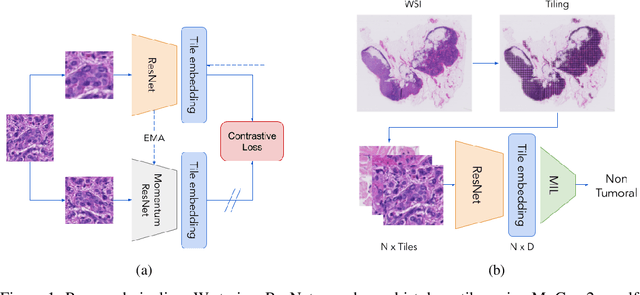
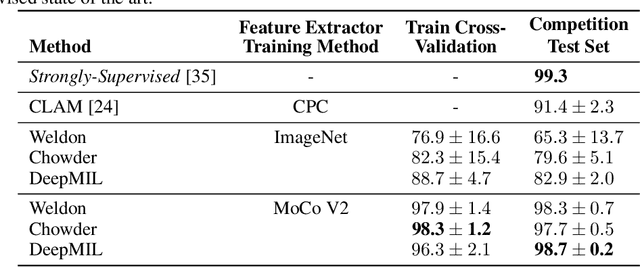
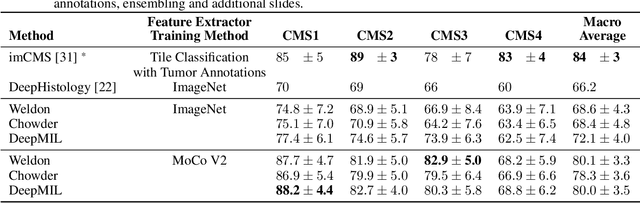
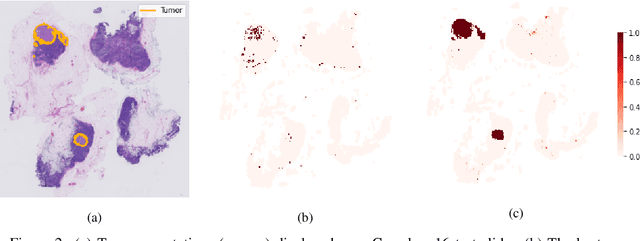
Abstract:One of the biggest challenges for applying machine learning to histopathology is weak supervision: whole-slide images have billions of pixels yet often only one global label. The state of the art therefore relies on strongly-supervised model training using additional local annotations from domain experts. However, in the absence of detailed annotations, most weakly-supervised approaches depend on a frozen feature extractor pre-trained on ImageNet. We identify this as a key weakness and propose to train an in-domain feature extractor on histology images using MoCo v2, a recent self-supervised learning algorithm. Experimental results on Camelyon16 and TCGA show that the proposed extractor greatly outperforms its ImageNet counterpart. In particular, our results improve the weakly-supervised state of the art on Camelyon16 from 91.4% to 98.7% AUC, thereby closing the gap with strongly-supervised models that reach 99.3% AUC. Through these experiments, we demonstrate that feature extractors trained via self-supervised learning can act as drop-in replacements to significantly improve existing machine learning techniques in histology. Lastly, we show that the learned embedding space exhibits biologically meaningful separation of tissue structures.
Classification and Disease Localization in Histopathology Using Only Global Labels: A Weakly-Supervised Approach
Feb 01, 2018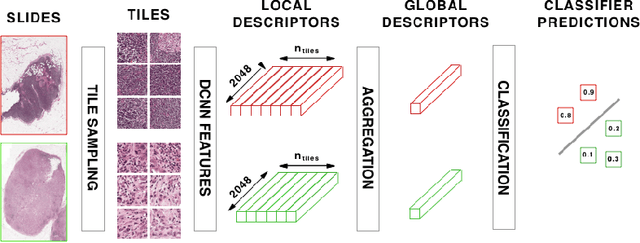
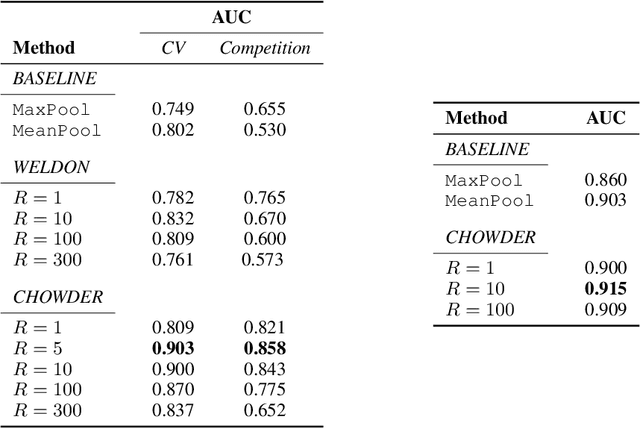
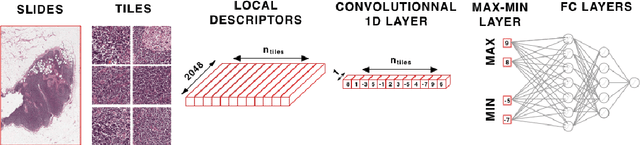
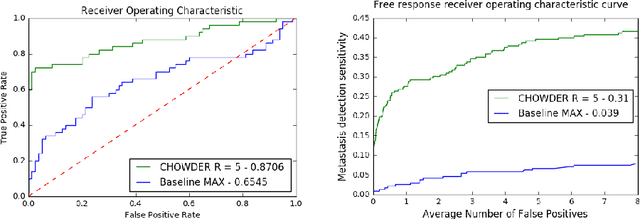
Abstract:Analysis of histopathology slides is a critical step for many diagnoses, and in particular in oncology where it defines the gold standard. In the case of digital histopathological analysis, highly trained pathologists must review vast whole-slide-images of extreme digital resolution ($100,000^2$ pixels) across multiple zoom levels in order to locate abnormal regions of cells, or in some cases single cells, out of millions. The application of deep learning to this problem is hampered not only by small sample sizes, as typical datasets contain only a few hundred samples, but also by the generation of ground-truth localized annotations for training interpretable classification and segmentation models. We propose a method for disease localization in the context of weakly supervised learning, where only image-level labels are available during training. Even without pixel-level annotations, we are able to demonstrate performance comparable with models trained with strong annotations on the Camelyon-16 lymph node metastases detection challenge. We accomplish this through the use of pre-trained deep convolutional networks, feature embedding, as well as learning via top instances and negative evidence, a multiple instance learning technique from the field of semantic segmentation and object detection.
Robust Detection of Covariate-Treatment Interactions in Clinical Trials
Dec 21, 2017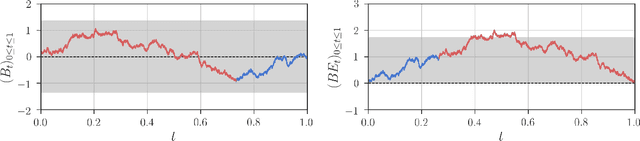

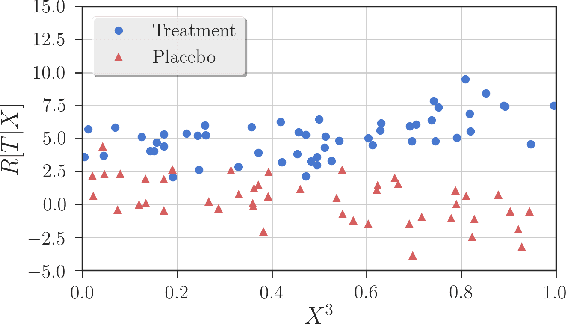
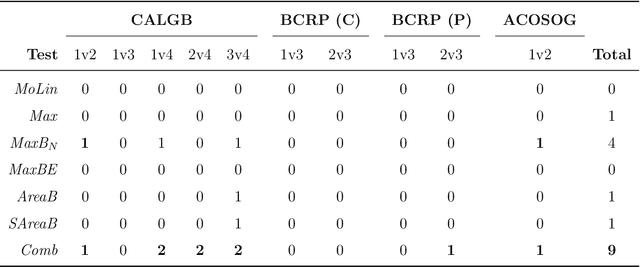
Abstract:Detection of interactions between treatment effects and patient descriptors in clinical trials is critical for optimizing the drug development process. The increasing volume of data accumulated in clinical trials provides a unique opportunity to discover new biomarkers and further the goal of personalized medicine, but it also requires innovative robust biomarker detection methods capable of detecting non-linear, and sometimes weak, signals. We propose a set of novel univariate statistical tests, based on the theory of random walks, which are able to capture non-linear and non-monotonic covariate-treatment interactions. We also propose a novel combined test, which leverages the power of all of our proposed univariate tests into a single general-case tool. We present results for both synthetic trials as well as real-world clinical trials, where we compare our method with state-of-the-art techniques and demonstrate the utility and robustness of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge