Olivier Moindrot
Self supervised learning improves dMMR/MSI detection from histology slides across multiple cancers
Sep 13, 2021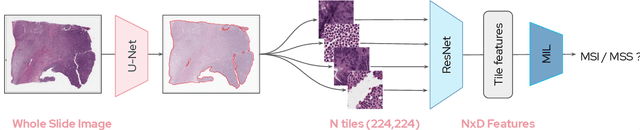

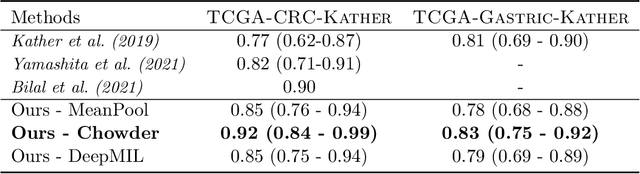
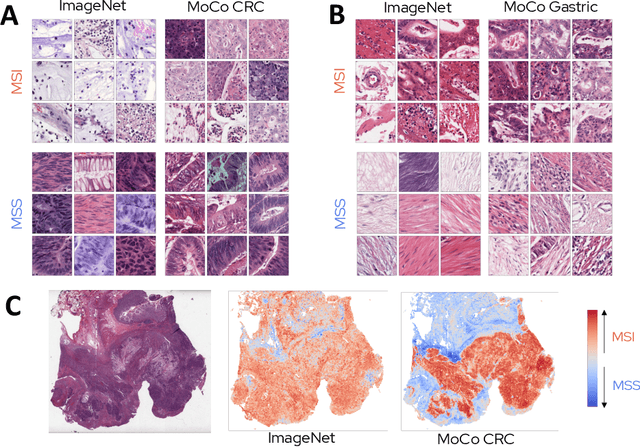
Abstract:Microsatellite instability (MSI) is a tumor phenotype whose diagnosis largely impacts patient care in colorectal cancers (CRC), and is associated with response to immunotherapy in all solid tumors. Deep learning models detecting MSI tumors directly from H&E stained slides have shown promise in improving diagnosis of MSI patients. Prior deep learning models for MSI detection have relied on neural networks pretrained on ImageNet dataset, which does not contain any medical image. In this study, we leverage recent advances in self-supervised learning by training neural networks on histology images from the TCGA dataset using MoCo V2. We show that these networks consistently outperform their counterparts pretrained using ImageNet and obtain state-of-the-art results for MSI detection with AUCs of 0.92 and 0.83 for CRC and gastric tumors, respectively. These models generalize well on an external CRC cohort (0.97 AUC on PAIP) and improve transfer from one organ to another. Finally we show that predictive image regions exhibit meaningful histological patterns, and that the use of MoCo features highlighted more relevant patterns according to an expert pathologist.
Using StyleGAN for Visual Interpretability of Deep Learning Models on Medical Images
Jan 19, 2021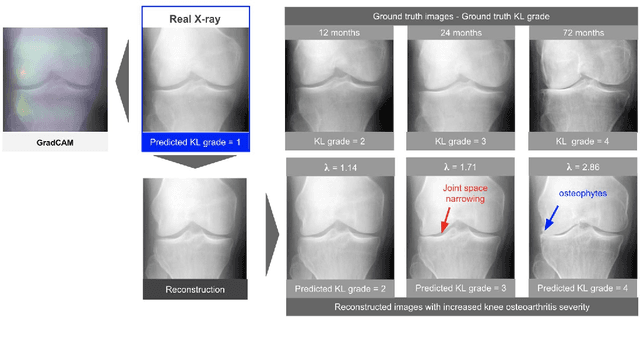
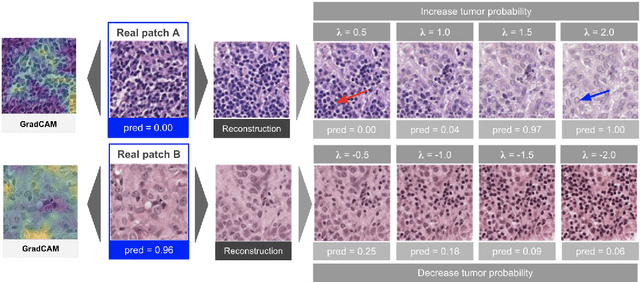
Abstract:As AI-based medical devices are becoming more common in imaging fields like radiology and histology, interpretability of the underlying predictive models is crucial to expand their use in clinical practice. Existing heatmap-based interpretability methods such as GradCAM only highlight the location of predictive features but do not explain how they contribute to the prediction. In this paper, we propose a new interpretability method that can be used to understand the predictions of any black-box model on images, by showing how the input image would be modified in order to produce different predictions. A StyleGAN is trained on medical images to provide a mapping between latent vectors and images. Our method identifies the optimal direction in the latent space to create a change in the model prediction. By shifting the latent representation of an input image along this direction, we can produce a series of new synthetic images with changed predictions. We validate our approach on histology and radiology images, and demonstrate its ability to provide meaningful explanations that are more informative than GradCAM heatmaps. Our method reveals the patterns learned by the model, which allows clinicians to build trust in the model's predictions, discover new biomarkers and eventually reveal potential biases.
Self-Supervision Closes the Gap Between Weak and Strong Supervision in Histology
Dec 07, 2020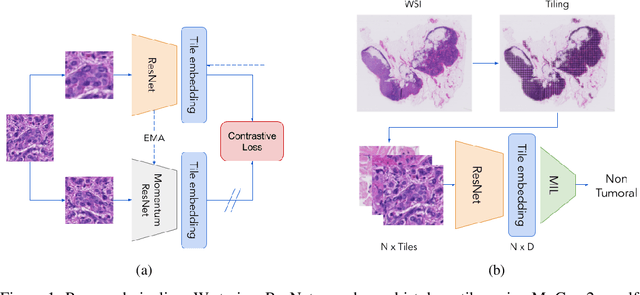
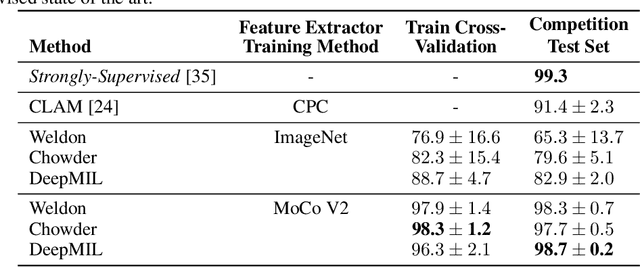
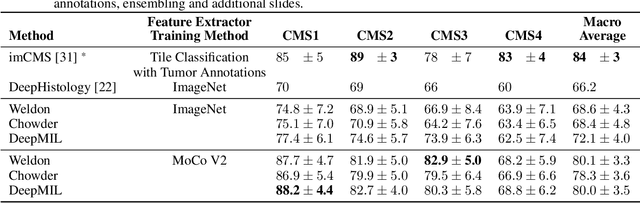
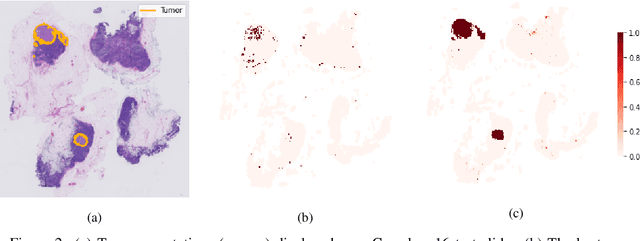
Abstract:One of the biggest challenges for applying machine learning to histopathology is weak supervision: whole-slide images have billions of pixels yet often only one global label. The state of the art therefore relies on strongly-supervised model training using additional local annotations from domain experts. However, in the absence of detailed annotations, most weakly-supervised approaches depend on a frozen feature extractor pre-trained on ImageNet. We identify this as a key weakness and propose to train an in-domain feature extractor on histology images using MoCo v2, a recent self-supervised learning algorithm. Experimental results on Camelyon16 and TCGA show that the proposed extractor greatly outperforms its ImageNet counterpart. In particular, our results improve the weakly-supervised state of the art on Camelyon16 from 91.4% to 98.7% AUC, thereby closing the gap with strongly-supervised models that reach 99.3% AUC. Through these experiments, we demonstrate that feature extractors trained via self-supervised learning can act as drop-in replacements to significantly improve existing machine learning techniques in histology. Lastly, we show that the learned embedding space exhibits biologically meaningful separation of tissue structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge