Benoit Schmauch
CARMIL: Context-Aware Regularization on Multiple Instance Learning models for Whole Slide Images
Aug 01, 2024



Abstract:Multiple Instance Learning (MIL) models have proven effective for cancer prognosis from Whole Slide Images. However, the original MIL formulation incorrectly assumes the patches of the same image to be independent, leading to a loss of spatial context as information flows through the network. Incorporating contextual knowledge into predictions is particularly important given the inclination for cancerous cells to form clusters and the presence of spatial indicators for tumors. State-of-the-art methods often use attention mechanisms eventually combined with graphs to capture spatial knowledge. In this paper, we take a novel and transversal approach, addressing this issue through the lens of regularization. We propose Context-Aware Regularization for Multiple Instance Learning (CARMIL), a versatile regularization scheme designed to seamlessly integrate spatial knowledge into any MIL model. Additionally, we present a new and generic metric to quantify the Context-Awareness of any MIL model when applied to Whole Slide Images, resolving a previously unexplored gap in the field. The efficacy of our framework is evaluated for two survival analysis tasks on glioblastoma (TCGA GBM) and colon cancer data (TCGA COAD).
Self supervised learning improves dMMR/MSI detection from histology slides across multiple cancers
Sep 13, 2021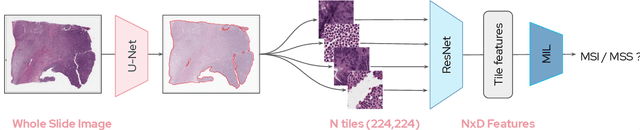

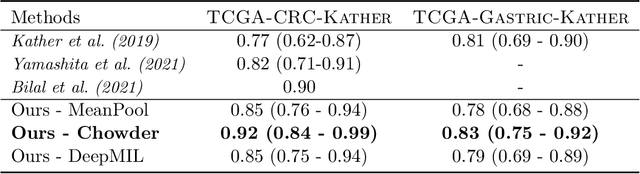
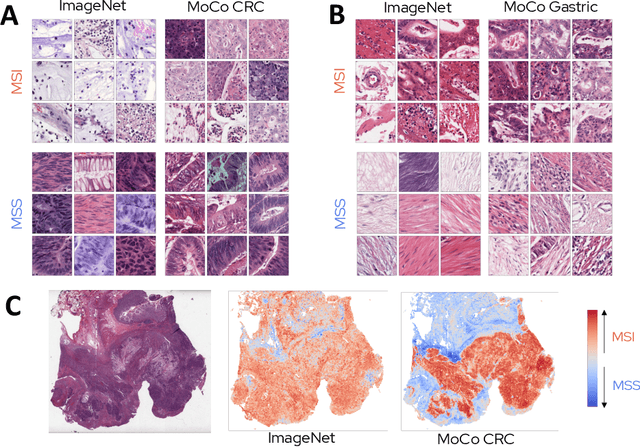
Abstract:Microsatellite instability (MSI) is a tumor phenotype whose diagnosis largely impacts patient care in colorectal cancers (CRC), and is associated with response to immunotherapy in all solid tumors. Deep learning models detecting MSI tumors directly from H&E stained slides have shown promise in improving diagnosis of MSI patients. Prior deep learning models for MSI detection have relied on neural networks pretrained on ImageNet dataset, which does not contain any medical image. In this study, we leverage recent advances in self-supervised learning by training neural networks on histology images from the TCGA dataset using MoCo V2. We show that these networks consistently outperform their counterparts pretrained using ImageNet and obtain state-of-the-art results for MSI detection with AUCs of 0.92 and 0.83 for CRC and gastric tumors, respectively. These models generalize well on an external CRC cohort (0.97 AUC on PAIP) and improve transfer from one organ to another. Finally we show that predictive image regions exhibit meaningful histological patterns, and that the use of MoCo features highlighted more relevant patterns according to an expert pathologist.
CNN+LSTM Architecture for Speech Emotion Recognition with Data Augmentation
Sep 11, 2018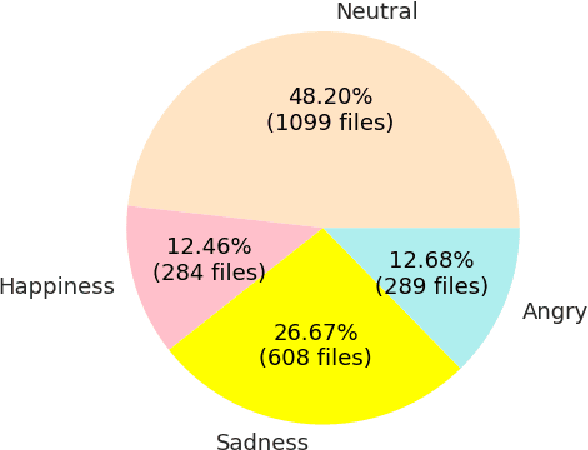

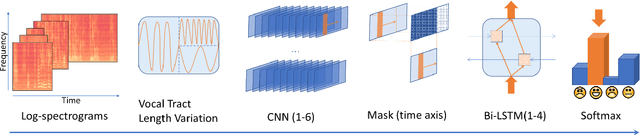
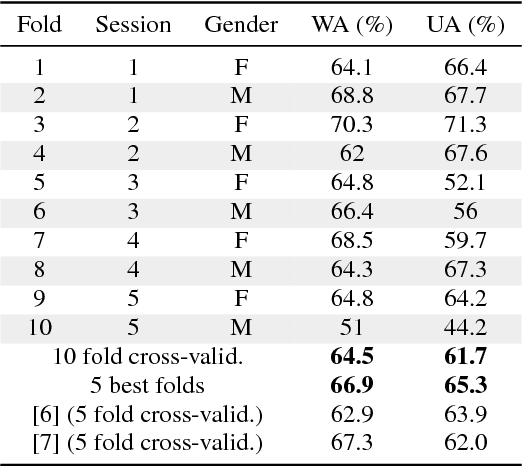
Abstract:In this work we design a neural network for recognizing emotions in speech, using the IEMOCAP dataset. Following the latest advances in audio analysis, we use an architecture involving both convolutional layers, for extracting high-level features from raw spectrograms, and recurrent ones for aggregating long-term dependencies. We examine the techniques of data augmentation with vocal track length perturbation, layer-wise optimizer adjustment, batch normalization of recurrent layers and obtain highly competitive results of 64.5% for weighted accuracy and 61.7% for unweighted accuracy on four emotions.
* 5 pages, 3 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge