Matthew Thomson
Automated mapping of virtual environments with visual predictive coding
Aug 20, 2023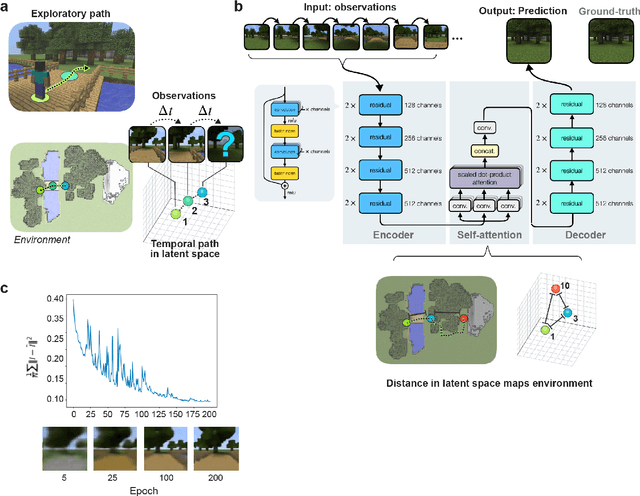
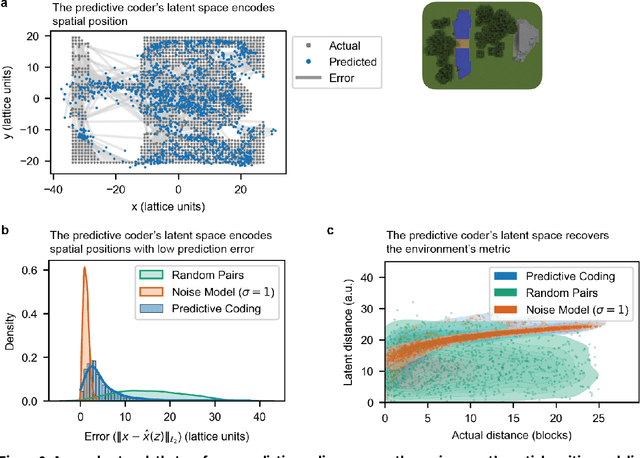
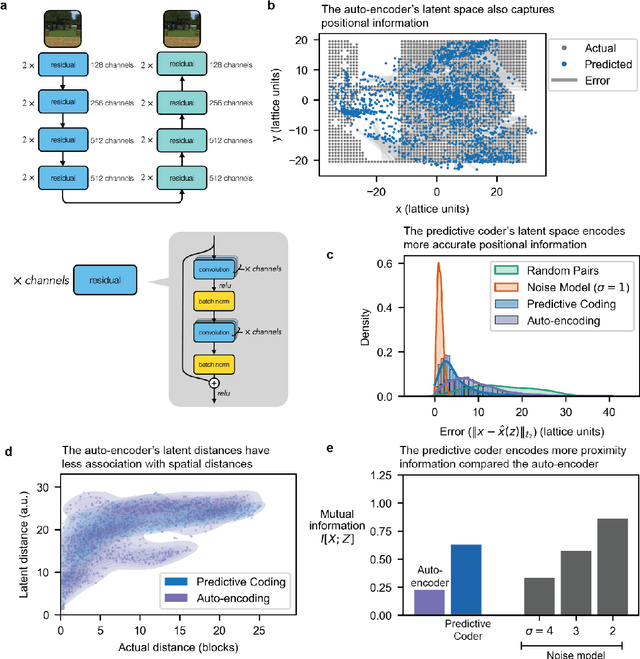
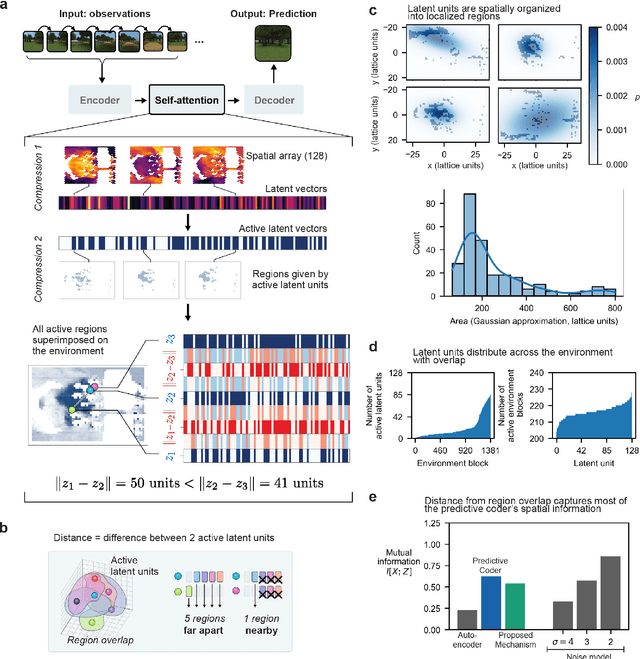
Abstract:Humans construct internal cognitive maps of their environment directly from sensory inputs without access to a system of explicit coordinates or distance measurements. While machine learning algorithms like SLAM utilize specialized visual inference procedures to identify visual features and construct spatial maps from visual and odometry data, the general nature of cognitive maps in the brain suggests a unified mapping algorithmic strategy that can generalize to auditory, tactile, and linguistic inputs. Here, we demonstrate that predictive coding provides a natural and versatile neural network algorithm for constructing spatial maps using sensory data. We introduce a framework in which an agent navigates a virtual environment while engaging in visual predictive coding using a self-attention-equipped convolutional neural network. While learning a next image prediction task, the agent automatically constructs an internal representation of the environment that quantitatively reflects distances. The internal map enables the agent to pinpoint its location relative to landmarks using only visual information.The predictive coding network generates a vectorized encoding of the environment that supports vector navigation where individual latent space units delineate localized, overlapping neighborhoods in the environment. Broadly, our work introduces predictive coding as a unified algorithmic framework for constructing cognitive maps that can naturally extend to the mapping of auditory, sensorimotor, and linguistic inputs.
CloudPred: Predicting Patient Phenotypes From Single-cell RNA-seq
Oct 13, 2021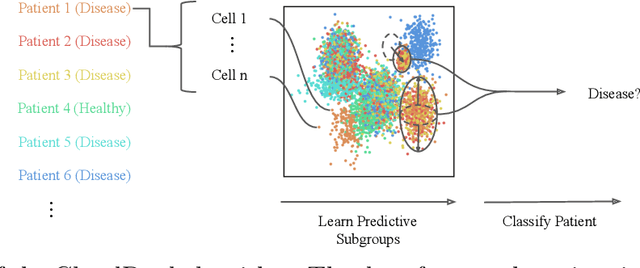
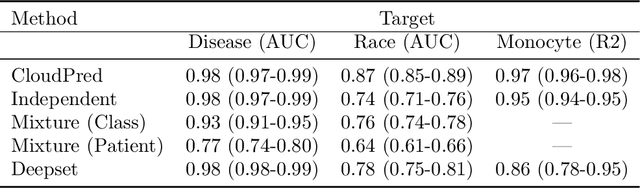
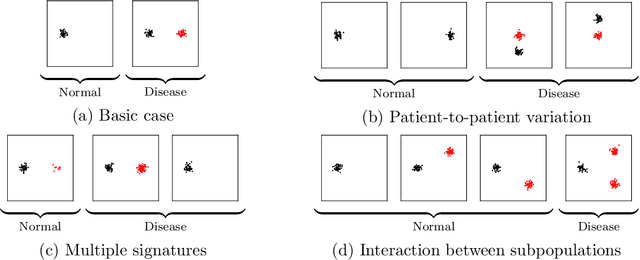
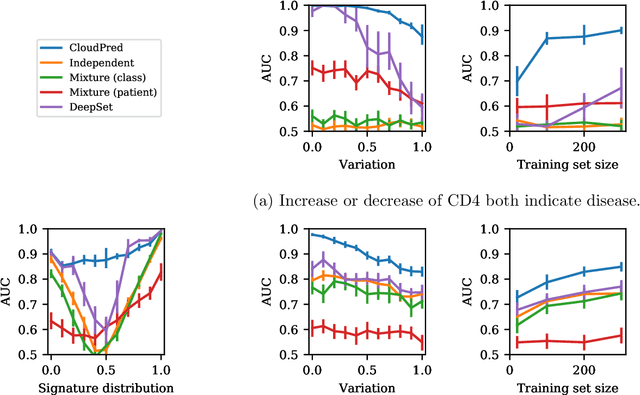
Abstract:Single-cell RNA sequencing (scRNA-seq) has the potential to provide powerful, high-resolution signatures to inform disease prognosis and precision medicine. This paper takes an important first step towards this goal by developing an interpretable machine learning algorithm, CloudPred, to predict individuals' disease phenotypes from their scRNA-seq data. Predicting phenotype from scRNA-seq is challenging for standard machine learning methods -- the number of cells measured can vary by orders of magnitude across individuals and the cell populations are also highly heterogeneous. Typical analysis creates pseudo-bulk samples which are biased toward prior annotations and also lose the single cell resolution. CloudPred addresses these challenges via a novel end-to-end differentiable learning algorithm which is coupled with a biologically informed mixture of cell types model. CloudPred automatically infers the cell subpopulation that are salient for the phenotype without prior annotations. We developed a systematic simulation platform to evaluate the performance of CloudPred and several alternative methods we propose, and find that CloudPred outperforms the alternative methods across several settings. We further validated CloudPred on a real scRNA-seq dataset of 142 lupus patients and controls. CloudPred achieves AUROC of 0.98 while identifying a specific subpopulation of CD4 T cells whose presence is highly indicative of lupus. CloudPred is a powerful new framework to predict clinical phenotypes from scRNA-seq data and to identify relevant cells.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge