James Gornet
Automated mapping of virtual environments with visual predictive coding
Aug 20, 2023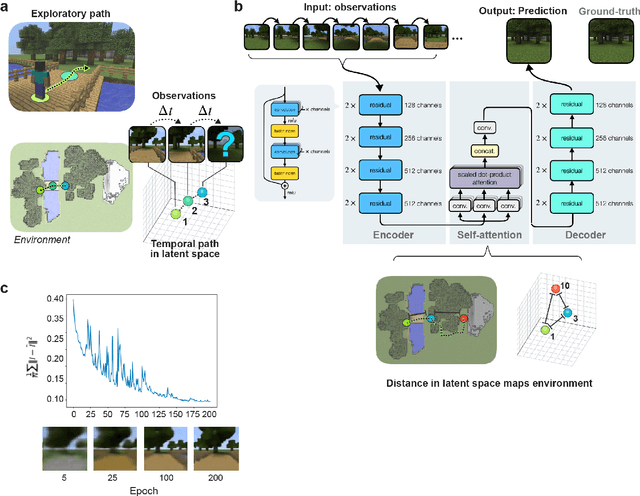
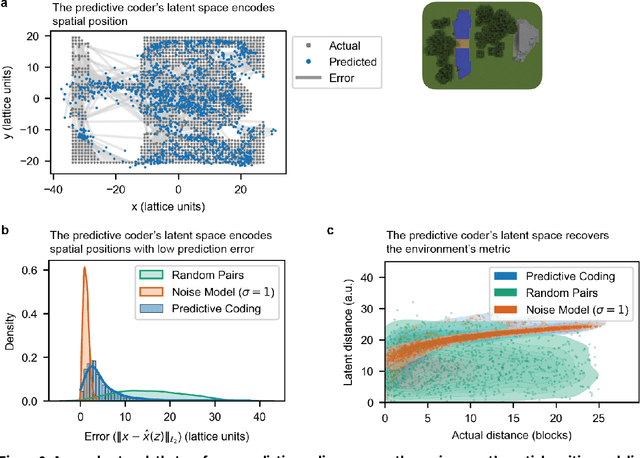
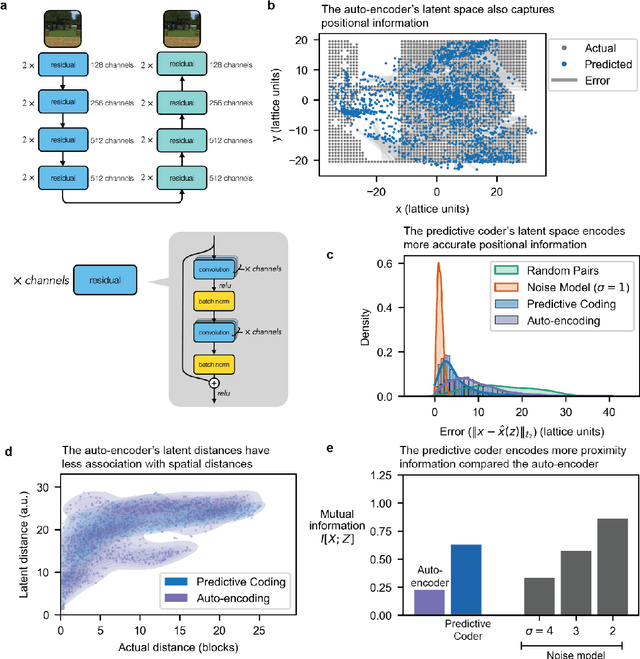
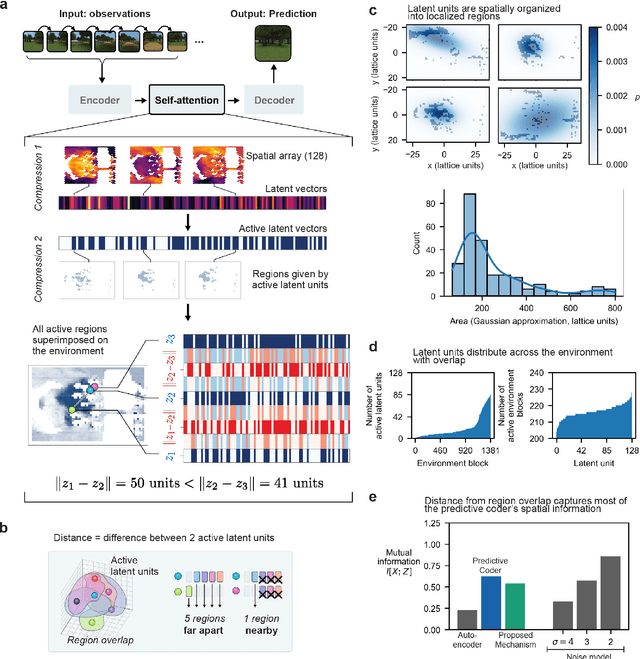
Abstract:Humans construct internal cognitive maps of their environment directly from sensory inputs without access to a system of explicit coordinates or distance measurements. While machine learning algorithms like SLAM utilize specialized visual inference procedures to identify visual features and construct spatial maps from visual and odometry data, the general nature of cognitive maps in the brain suggests a unified mapping algorithmic strategy that can generalize to auditory, tactile, and linguistic inputs. Here, we demonstrate that predictive coding provides a natural and versatile neural network algorithm for constructing spatial maps using sensory data. We introduce a framework in which an agent navigates a virtual environment while engaging in visual predictive coding using a self-attention-equipped convolutional neural network. While learning a next image prediction task, the agent automatically constructs an internal representation of the environment that quantitatively reflects distances. The internal map enables the agent to pinpoint its location relative to landmarks using only visual information.The predictive coding network generates a vectorized encoding of the environment that supports vector navigation where individual latent space units delineate localized, overlapping neighborhoods in the environment. Broadly, our work introduces predictive coding as a unified algorithmic framework for constructing cognitive maps that can naturally extend to the mapping of auditory, sensorimotor, and linguistic inputs.
Fine-Grained System Identification of Nonlinear Neural Circuits
Jun 09, 2021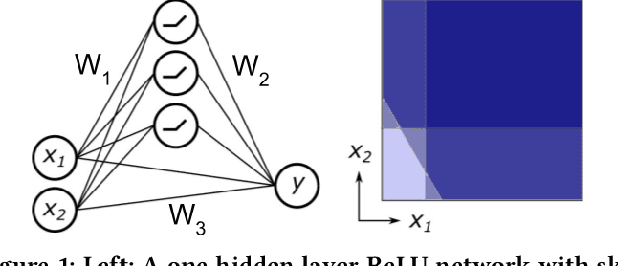
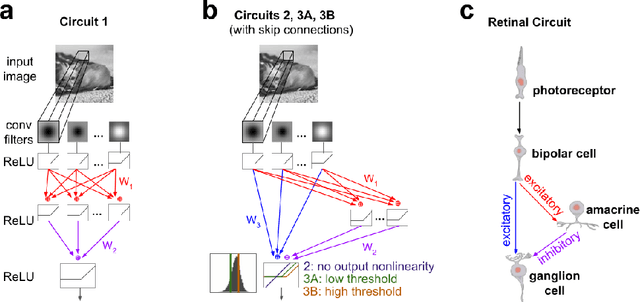
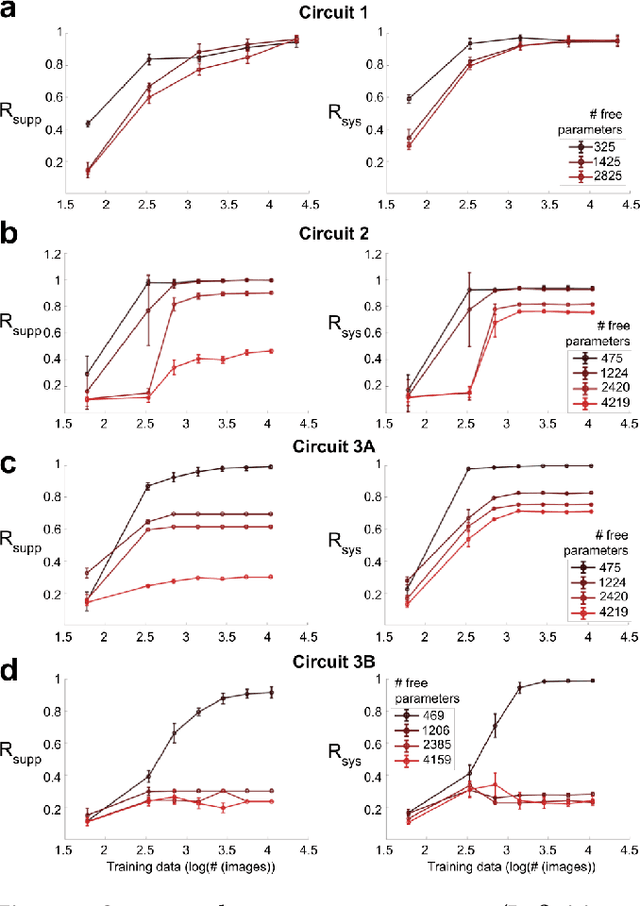
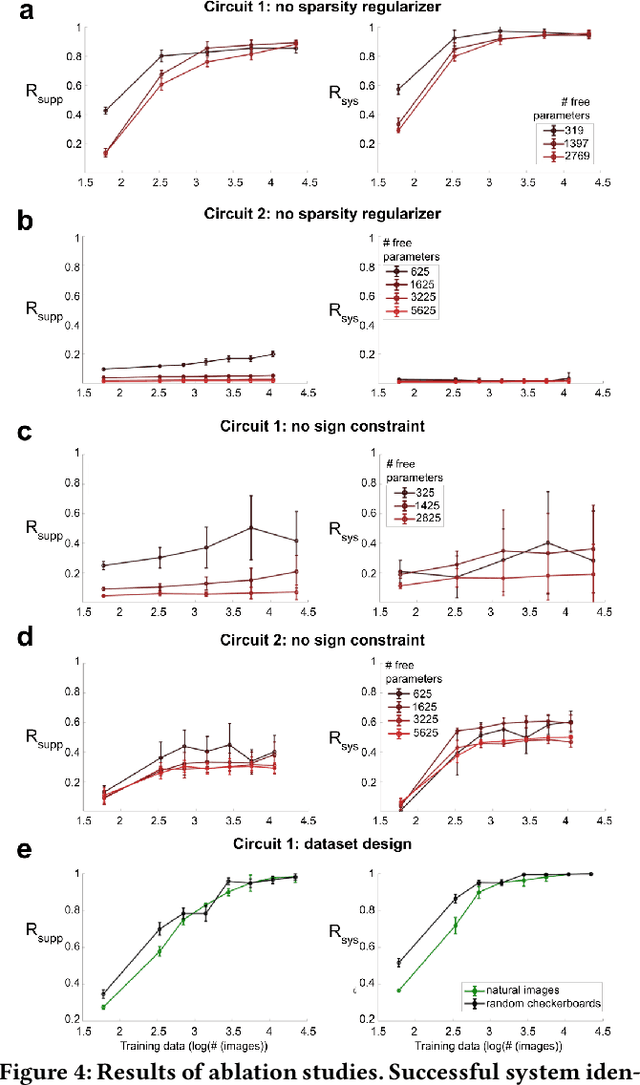
Abstract:We study the problem of sparse nonlinear model recovery of high dimensional compositional functions. Our study is motivated by emerging opportunities in neuroscience to recover fine-grained models of biological neural circuits using collected measurement data. Guided by available domain knowledge in neuroscience, we explore conditions under which one can recover the underlying biological circuit that generated the training data. Our results suggest insights of both theoretical and practical interests. Most notably, we find that a sign constraint on the weights is a necessary condition for system recovery, which we establish both theoretically with an identifiability guarantee and empirically on simulated biological circuits. We conclude with a case study on retinal ganglion cell circuits using data collected from mouse retina, showcasing the practical potential of this approach.
Neural Networks with Recurrent Generative Feedback
Jul 17, 2020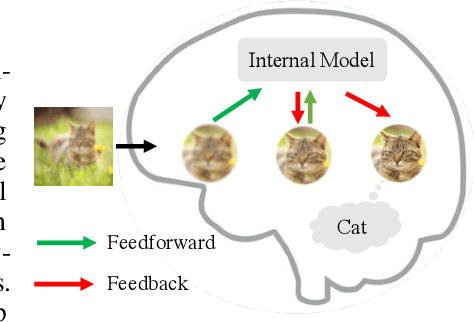
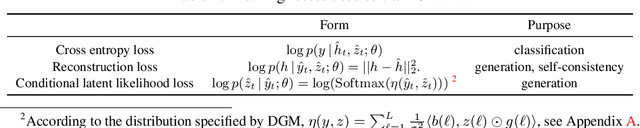
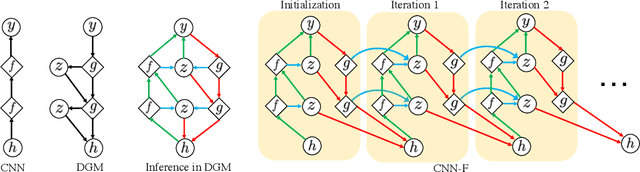
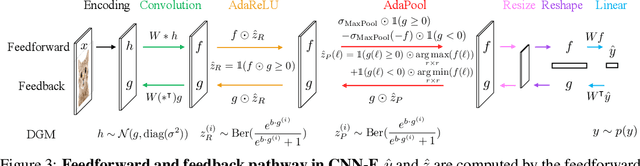
Abstract:Neural networks are vulnerable to input perturbations such as additive noise and adversarial attacks. In contrast, human perception is much more robust to such perturbations. The Bayesian brain hypothesis states that human brains use an internal generative model to update the posterior beliefs of the sensory input. This mechanism can be interpreted as a form of self-consistency between the maximum a posteriori (MAP) estimation of the internal generative model and the external environmental. Inspired by this, we enforce consistency in neural networks by incorporating generative recurrent feedback. We instantiate it on convolutional neural networks (CNNs). The proposed framework, termed Convolutional Neural Networks with Feedback (CNN-F), introduces a generative feedback with latent variables into existing CNN architectures, making consistent predictions via alternating MAP inference under a Bayesian framework. CNN-F shows considerably better adversarial robustness over regular feedforward CNNs on standard benchmarks. In addition, With higher V4 and IT neural predictivity, CNN-F produces object representations closer to primate vision than conventional CNNs.
Reconstructing neuronal anatomy from whole-brain images
Mar 17, 2019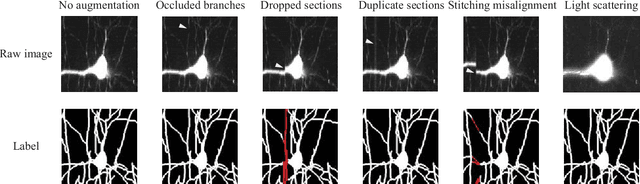
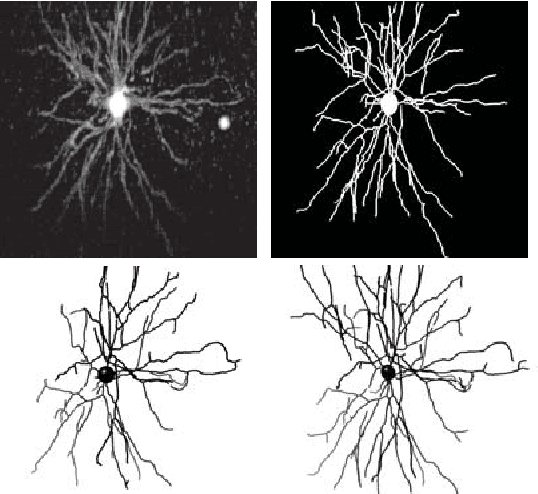
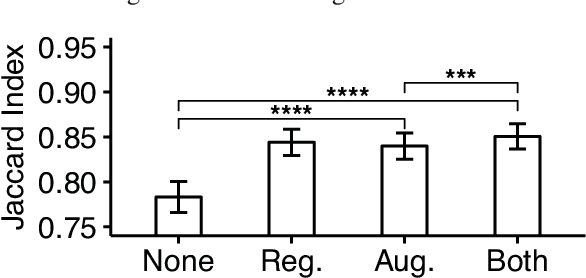
Abstract:Reconstructing multiple molecularly defined neurons from individual brains and across multiple brain regions can reveal organizational principles of the nervous system. However, high resolution imaging of the whole brain is a technically challenging and slow process. Recently, oblique light sheet microscopy has emerged as a rapid imaging method that can provide whole brain fluorescence microscopy at a voxel size of 0.4 by 0.4 by 2.5 cubic microns. On the other hand, complex image artifacts due to whole-brain coverage produce apparent discontinuities in neuronal arbors. Here, we present connectivity-preserving methods and data augmentation strategies for supervised learning of neuroanatomy from light microscopy using neural networks. We quantify the merit of our approach by implementing an end-to-end automated tracing pipeline. Lastly, we demonstrate a scalable, distributed implementation that can reconstruct the large datasets that sub-micron whole-brain images produce.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge