Anima Anandkumar
Decoupled Diffusion Sampling for Inverse Problems on Function Spaces
Jan 30, 2026Abstract:We propose a data-efficient, physics-aware generative framework in function space for inverse PDE problems. Existing plug-and-play diffusion posterior samplers represent physics implicitly through joint coefficient-solution modeling, requiring substantial paired supervision. In contrast, our Decoupled Diffusion Inverse Solver (DDIS) employs a decoupled design: an unconditional diffusion learns the coefficient prior, while a neural operator explicitly models the forward PDE for guidance. This decoupling enables superior data efficiency and effective physics-informed learning, while naturally supporting Decoupled Annealing Posterior Sampling (DAPS) to avoid over-smoothing in Diffusion Posterior Sampling (DPS). Theoretically, we prove that DDIS avoids the guidance attenuation failure of joint models when training data is scarce. Empirically, DDIS achieves state-of-the-art performance under sparse observation, improving $l_2$ error by 11% and spectral error by 54% on average; when data is limited to 1%, DDIS maintains accuracy with 40% advantage in $l_2$ error compared to joint models.
Resolution-Independent Neural Operators for Multi-Rate Sparse-View CT
Dec 13, 2025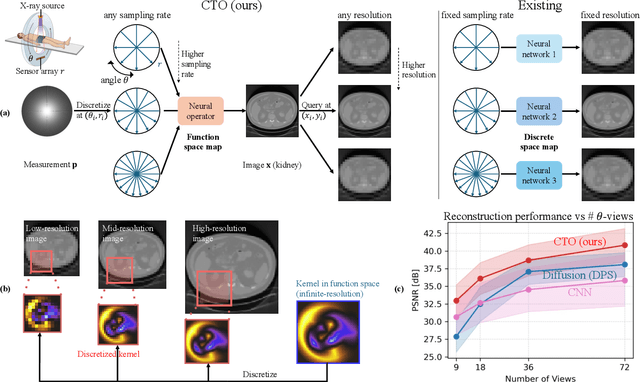
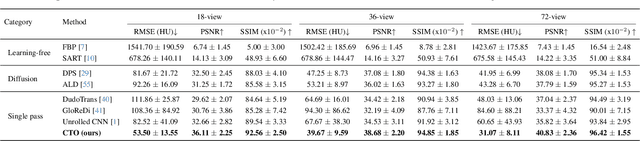
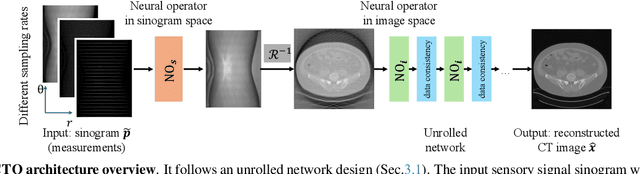
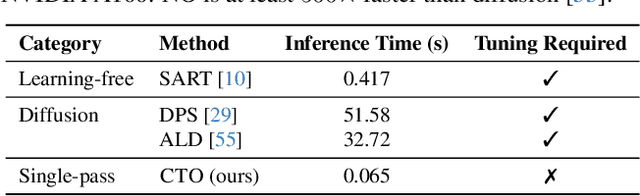
Abstract:Sparse-view Computed Tomography (CT) reconstructs images from a limited number of X-ray projections to reduce radiation and scanning time, which makes reconstruction an ill-posed inverse problem. Deep learning methods achieve high-fidelity reconstructions but often overfit to a fixed acquisition setup, failing to generalize across sampling rates and image resolutions. For example, convolutional neural networks (CNNs) use the same learned kernels across resolutions, leading to artifacts when data resolution changes. We propose Computed Tomography neural Operator (CTO), a unified CT reconstruction framework that extends to continuous function space, enabling generalization (without retraining) across sampling rates and image resolutions. CTO operates jointly in the sinogram and image domains through rotation-equivariant Discrete-Continuous convolutions parametrized in the function space, making it inherently resolution- and sampling-agnostic. Empirically, CTO enables consistent multi-sampling-rate and cross-resolution performance, with on average >4dB PSNR gain over CNNs. Compared to state-of-the-art diffusion methods, CTO is 500$\times$ faster in inference time with on average 3dB gain. Empirical results also validate our design choices behind CTO's sinogram-space operator learning and rotation-equivariant convolution. Overall, CTO outperforms state-of-the-art baselines across sampling rates and resolutions, offering a scalable and generalizable solution that makes automated CT reconstruction more practical for deployment.
Self Distillation Fine-Tuning of Protein Language Models Improves Versatility in Protein Design
Dec 10, 2025Abstract:Supervised fine-tuning (SFT) is a standard approach for adapting large language models to specialized domains, yet its application to protein sequence modeling and protein language models (PLMs) remains ad hoc. This is in part because high-quality annotated data are far more difficult to obtain for proteins than for natural language. We present a simple and general recipe for fast SFT of PLMs, designed to improve the fidelity, reliability, and novelty of generated protein sequences. Unlike existing approaches that require costly precompiled experimental datasets for SFT, our method leverages the PLM itself, integrating a lightweight curation pipeline with domain-specific filters to construct high-quality training data. These filters can independently refine a PLM's output and identify candidates for in vitro evaluation; when combined with SFT, they enable PLMs to generate more stable and functional enzymes, while expanding exploration into protein sequence space beyond natural variants. Although our approach is agnostic to both the choice of protein language model (PLM) and the protein system, we demonstrate its effectiveness with a genome-scale PLM (GenSLM) applied to the tryptophan synthase enzyme family. The supervised fine-tuned model generates sequences that are not only more novel but also display improved characteristics across both targeted design constraints and emergent protein property measures.
Generating Natural-Language Surgical Feedback: From Structured Representation to Domain-Grounded Evaluation
Nov 19, 2025Abstract:High-quality intraoperative feedback from a surgical trainer is pivotal for improving trainee performance and long-term skill acquisition. Automating natural, trainer-style feedback promises timely, accessible, and consistent guidance at scale but requires models that understand clinically relevant representations. We present a structure-aware pipeline that learns a surgical action ontology from real trainer-to-trainee transcripts (33 surgeries) and uses it to condition feedback generation. We contribute by (1) mining Instrument-Action-Target (IAT) triplets from real-world feedback text and clustering surface forms into normalized categories, (2) fine-tuning a video-to-IAT model that leverages the surgical procedure and task contexts as well as fine-grained temporal instrument motion, and (3) demonstrating how to effectively use IAT triplet representations to guide GPT-4o in generating clinically grounded, trainer-style feedback. We show that, on Task 1: Video-to-IAT recognition, our context injection and temporal tracking deliver consistent AUC gains (Instrument: 0.67 to 0.74; Action: 0.60 to 0.63; Tissue: 0.74 to 0.79). For Task 2: feedback text generation (rated on a 1-5 fidelity rubric where 1 = opposite/unsafe, 3 = admissible, and 5 = perfect match to a human trainer), GPT-4o from video alone scores 2.17, while IAT conditioning reaches 2.44 (+12.4%), doubling the share of admissible generations with score >= 3 from 21% to 42%. Traditional text-similarity metrics also improve: word error rate decreases by 15-31% and ROUGE (phrase/substring overlap) increases by 9-64%. Grounding generation in explicit IAT structure improves fidelity and yields clinician-verifiable rationales, supporting auditable use in surgical training.
EcoSpa: Efficient Transformer Training with Coupled Sparsity
Nov 09, 2025Abstract:Transformers have become the backbone of modern AI, yet their high computational demands pose critical system challenges. While sparse training offers efficiency gains, existing methods fail to preserve critical structural relationships between weight matrices that interact multiplicatively in attention and feed-forward layers. This oversight leads to performance degradation at high sparsity levels. We introduce EcoSpa, an efficient structured sparse training method that jointly evaluates and sparsifies coupled weight matrix pairs, preserving their interaction patterns through aligned row/column removal. EcoSpa introduces a new granularity for calibrating structural component importance and performs coupled estimation and sparsification across both pre-training and fine-tuning scenarios. Evaluations demonstrate substantial improvements: EcoSpa enables efficient training of LLaMA-1B with 50\% memory reduction and 21\% faster training, achieves $2.2\times$ model compression on GPT-2-Medium with $2.4$ lower perplexity, and delivers $1.6\times$ inference speedup. The approach uses standard PyTorch operations, requiring no custom hardware or kernels, making efficient transformer training accessible on commodity hardware.
The Personality Illusion: Revealing Dissociation Between Self-Reports & Behavior in LLMs
Sep 03, 2025Abstract:Personality traits have long been studied as predictors of human behavior.Recent advances in Large Language Models (LLMs) suggest similar patterns may emerge in artificial systems, with advanced LLMs displaying consistent behavioral tendencies resembling human traits like agreeableness and self-regulation. Understanding these patterns is crucial, yet prior work primarily relied on simplified self-reports and heuristic prompting, with little behavioral validation. In this study, we systematically characterize LLM personality across three dimensions: (1) the dynamic emergence and evolution of trait profiles throughout training stages; (2) the predictive validity of self-reported traits in behavioral tasks; and (3) the impact of targeted interventions, such as persona injection, on both self-reports and behavior. Our findings reveal that instructional alignment (e.g., RLHF, instruction tuning) significantly stabilizes trait expression and strengthens trait correlations in ways that mirror human data. However, these self-reported traits do not reliably predict behavior, and observed associations often diverge from human patterns. While persona injection successfully steers self-reports in the intended direction, it exerts little or inconsistent effect on actual behavior. By distinguishing surface-level trait expression from behavioral consistency, our findings challenge assumptions about LLM personality and underscore the need for deeper evaluation in alignment and interpretability.
Sparse Autoencoder Neural Operators: Model Recovery in Function Spaces
Sep 03, 2025Abstract:We frame the problem of unifying representations in neural models as one of sparse model recovery and introduce a framework that extends sparse autoencoders (SAEs) to lifted spaces and infinite-dimensional function spaces, enabling mechanistic interpretability of large neural operators (NO). While the Platonic Representation Hypothesis suggests that neural networks converge to similar representations across architectures, the representational properties of neural operators remain underexplored despite their growing importance in scientific computing. We compare the inference and training dynamics of SAEs, lifted-SAE, and SAE neural operators. We highlight how lifting and operator modules introduce beneficial inductive biases, enabling faster recovery, improved recovery of smooth concepts, and robust inference across varying resolutions, a property unique to neural operators.
Geometric Operator Learning with Optimal Transport
Jul 26, 2025Abstract:We propose integrating optimal transport (OT) into operator learning for partial differential equations (PDEs) on complex geometries. Classical geometric learning methods typically represent domains as meshes, graphs, or point clouds. Our approach generalizes discretized meshes to mesh density functions, formulating geometry embedding as an OT problem that maps these functions to a uniform density in a reference space. Compared to previous methods relying on interpolation or shared deformation, our OT-based method employs instance-dependent deformation, offering enhanced flexibility and effectiveness. For 3D simulations focused on surfaces, our OT-based neural operator embeds the surface geometry into a 2D parameterized latent space. By performing computations directly on this 2D representation of the surface manifold, it achieves significant computational efficiency gains compared to volumetric simulation. Experiments with Reynolds-averaged Navier-Stokes equations (RANS) on the ShapeNet-Car and DrivAerNet-Car datasets show that our method achieves better accuracy and also reduces computational expenses in terms of both time and memory usage compared to existing machine learning models. Additionally, our model demonstrates significantly improved accuracy on the FlowBench dataset, underscoring the benefits of employing instance-dependent deformation for datasets with highly variable geometries.
FourCastNet 3: A geometric approach to probabilistic machine-learning weather forecasting at scale
Jul 16, 2025Abstract:FourCastNet 3 advances global weather modeling by implementing a scalable, geometric machine learning (ML) approach to probabilistic ensemble forecasting. The approach is designed to respect spherical geometry and to accurately model the spatially correlated probabilistic nature of the problem, resulting in stable spectra and realistic dynamics across multiple scales. FourCastNet 3 delivers forecasting accuracy that surpasses leading conventional ensemble models and rivals the best diffusion-based methods, while producing forecasts 8 to 60 times faster than these approaches. In contrast to other ML approaches, FourCastNet 3 demonstrates excellent probabilistic calibration and retains realistic spectra, even at extended lead times of up to 60 days. All of these advances are realized using a purely convolutional neural network architecture tailored for spherical geometry. Scalable and efficient large-scale training on 1024 GPUs and more is enabled by a novel training paradigm for combined model- and data-parallelism, inspired by domain decomposition methods in classical numerical models. Additionally, FourCastNet 3 enables rapid inference on a single GPU, producing a 90-day global forecast at 0.25{\deg}, 6-hourly resolution in under 20 seconds. Its computational efficiency, medium-range probabilistic skill, spectral fidelity, and rollout stability at subseasonal timescales make it a strong candidate for improving meteorological forecasting and early warning systems through large ensemble predictions.
High precision PINNs in unbounded domains: application to singularity formulation in PDEs
Jun 24, 2025Abstract:We investigate the high-precision training of Physics-Informed Neural Networks (PINNs) in unbounded domains, with a special focus on applications to singularity formulation in PDEs. We propose a modularized approach and study the choices of neural network ansatz, sampling strategy, and optimization algorithm. When combined with rigorous computer-assisted proofs and PDE analysis, the numerical solutions identified by PINNs, provided they are of high precision, can serve as a powerful tool for studying singularities in PDEs. For 1D Burgers equation, our framework can lead to a solution with very high precision, and for the 2D Boussinesq equation, which is directly related to the singularity formulation in 3D Euler and Navier-Stokes equations, we obtain a solution whose loss is $4$ digits smaller than that obtained in \cite{wang2023asymptotic} with fewer training steps. We also discuss potential directions for pushing towards machine precision for higher-dimensional problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge