J. Gregory Pauloski
HiPerRAG: High-Performance Retrieval Augmented Generation for Scientific Insights
May 07, 2025Abstract:The volume of scientific literature is growing exponentially, leading to underutilized discoveries, duplicated efforts, and limited cross-disciplinary collaboration. Retrieval Augmented Generation (RAG) offers a way to assist scientists by improving the factuality of Large Language Models (LLMs) in processing this influx of information. However, scaling RAG to handle millions of articles introduces significant challenges, including the high computational costs associated with parsing documents and embedding scientific knowledge, as well as the algorithmic complexity of aligning these representations with the nuanced semantics of scientific content. To address these issues, we introduce HiPerRAG, a RAG workflow powered by high performance computing (HPC) to index and retrieve knowledge from more than 3.6 million scientific articles. At its core are Oreo, a high-throughput model for multimodal document parsing, and ColTrast, a query-aware encoder fine-tuning algorithm that enhances retrieval accuracy by using contrastive learning and late-interaction techniques. HiPerRAG delivers robust performance on existing scientific question answering benchmarks and two new benchmarks introduced in this work, achieving 90% accuracy on SciQ and 76% on PubMedQA-outperforming both domain-specific models like PubMedGPT and commercial LLMs such as GPT-4. Scaling to thousands of GPUs on the Polaris, Sunspot, and Frontier supercomputers, HiPerRAG delivers million document-scale RAG workflows for unifying scientific knowledge and fostering interdisciplinary innovation.
MOFA: Discovering Materials for Carbon Capture with a GenAI- and Simulation-Based Workflow
Jan 18, 2025Abstract:We present MOFA, an open-source generative AI (GenAI) plus simulation workflow for high-throughput generation of metal-organic frameworks (MOFs) on large-scale high-performance computing (HPC) systems. MOFA addresses key challenges in integrating GPU-accelerated computing for GPU-intensive GenAI tasks, including distributed training and inference, alongside CPU- and GPU-optimized tasks for screening and filtering AI-generated MOFs using molecular dynamics, density functional theory, and Monte Carlo simulations. These heterogeneous tasks are unified within an online learning framework that optimizes the utilization of available CPU and GPU resources across HPC systems. Performance metrics from a 450-node (14,400 AMD Zen 3 CPUs + 1800 NVIDIA A100 GPUs) supercomputer run demonstrate that MOFA achieves high-throughput generation of novel MOF structures, with CO$_2$ adsorption capacities ranking among the top 10 in the hypothetical MOF (hMOF) dataset. Furthermore, the production of high-quality MOFs exhibits a linear relationship with the number of nodes utilized. The modular architecture of MOFA will facilitate its integration into other scientific applications that dynamically combine GenAI with large-scale simulations.
Flight: A FaaS-Based Framework for Complex and Hierarchical Federated Learning
Sep 24, 2024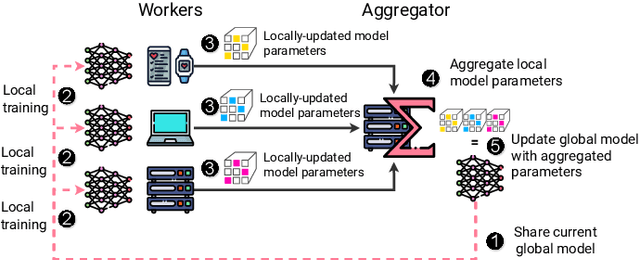
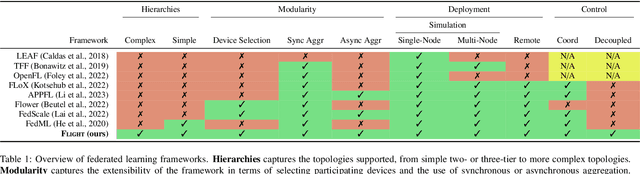
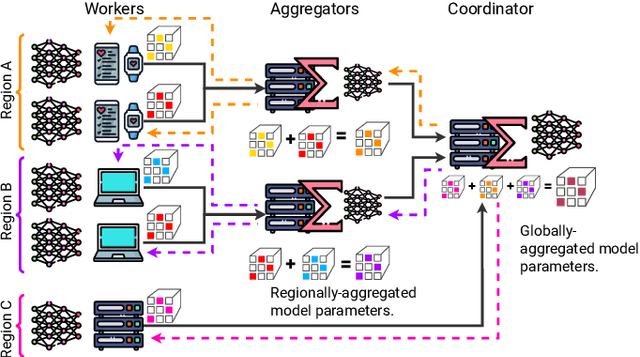
Abstract:Federated Learning (FL) is a decentralized machine learning paradigm where models are trained on distributed devices and are aggregated at a central server. Existing FL frameworks assume simple two-tier network topologies where end devices are directly connected to the aggregation server. While this is a practical mental model, it does not exploit the inherent topology of real-world distributed systems like the Internet-of-Things. We present Flight, a novel FL framework that supports complex hierarchical multi-tier topologies, asynchronous aggregation, and decouples the control plane from the data plane. We compare the performance of Flight against Flower, a state-of-the-art FL framework. Our results show that Flight scales beyond Flower, supporting up to 2048 simultaneous devices, and reduces FL makespan across several models. Finally, we show that Flight's hierarchical FL model can reduce communication overheads by more than 60%.
Employing Artificial Intelligence to Steer Exascale Workflows with Colmena
Aug 26, 2024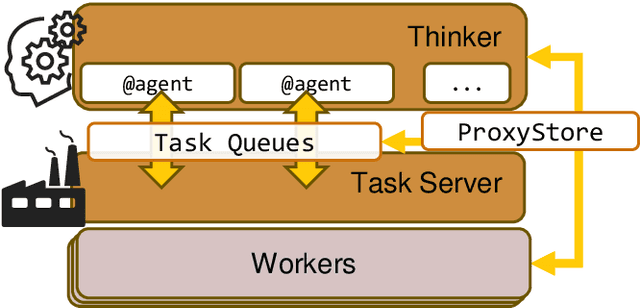
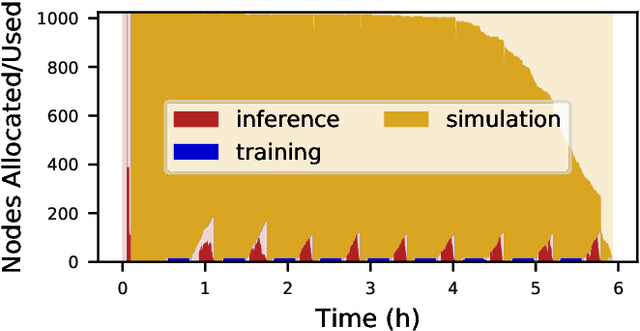
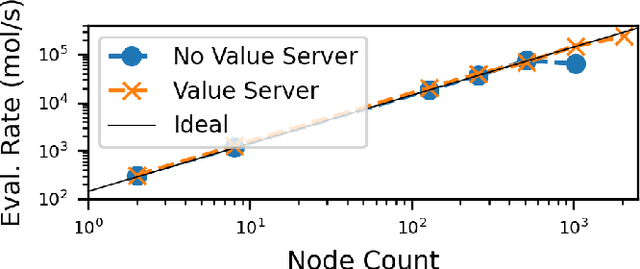
Abstract:Computational workflows are a common class of application on supercomputers, yet the loosely coupled and heterogeneous nature of workflows often fails to take full advantage of their capabilities. We created Colmena to leverage the massive parallelism of a supercomputer by using Artificial Intelligence (AI) to learn from and adapt a workflow as it executes. Colmena allows scientists to define how their application should respond to events (e.g., task completion) as a series of cooperative agents. In this paper, we describe the design of Colmena, the challenges we overcame while deploying applications on exascale systems, and the science workflows we have enhanced through interweaving AI. The scaling challenges we discuss include developing steering strategies that maximize node utilization, introducing data fabrics that reduce communication overhead of data-intensive tasks, and implementing workflow tasks that cache costly operations between invocations. These innovations coupled with a variety of application patterns accessible through our agent-based steering model have enabled science advances in chemistry, biophysics, and materials science using different types of AI. Our vision is that Colmena will spur creative solutions that harness AI across many domains of scientific computing.
Trillion Parameter AI Serving Infrastructure for Scientific Discovery: A Survey and Vision
Feb 05, 2024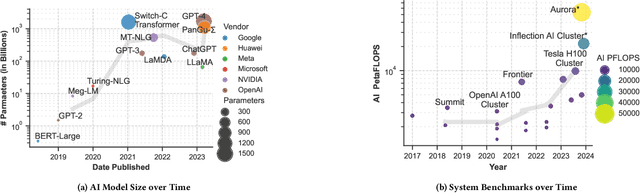
Abstract:Deep learning methods are transforming research, enabling new techniques, and ultimately leading to new discoveries. As the demand for more capable AI models continues to grow, we are now entering an era of Trillion Parameter Models (TPM), or models with more than a trillion parameters -- such as Huawei's PanGu-$\Sigma$. We describe a vision for the ecosystem of TPM users and providers that caters to the specific needs of the scientific community. We then outline the significant technical challenges and open problems in system design for serving TPMs to enable scientific research and discovery. Specifically, we describe the requirements of a comprehensive software stack and interfaces to support the diverse and flexible requirements of researchers.
DeepSpeed4Science Initiative: Enabling Large-Scale Scientific Discovery through Sophisticated AI System Technologies
Oct 11, 2023



Abstract:In the upcoming decade, deep learning may revolutionize the natural sciences, enhancing our capacity to model and predict natural occurrences. This could herald a new era of scientific exploration, bringing significant advancements across sectors from drug development to renewable energy. To answer this call, we present DeepSpeed4Science initiative (deepspeed4science.ai) which aims to build unique capabilities through AI system technology innovations to help domain experts to unlock today's biggest science mysteries. By leveraging DeepSpeed's current technology pillars (training, inference and compression) as base technology enablers, DeepSpeed4Science will create a new set of AI system technologies tailored for accelerating scientific discoveries by addressing their unique complexity beyond the common technical approaches used for accelerating generic large language models (LLMs). In this paper, we showcase the early progress we made with DeepSpeed4Science in addressing two of the critical system challenges in structural biology research.
Cloud Services Enable Efficient AI-Guided Simulation Workflows across Heterogeneous Resources
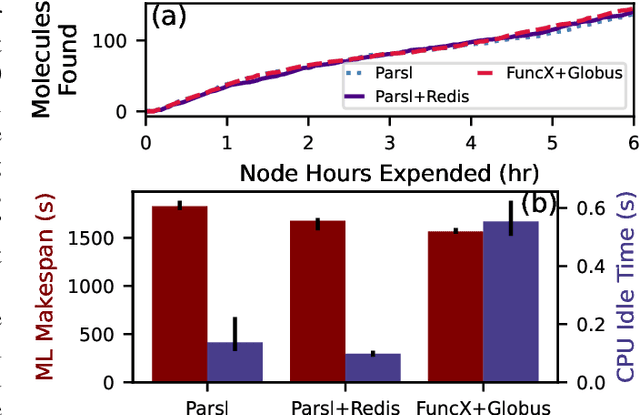
Mar 15, 2023Abstract:Applications that fuse machine learning and simulation can benefit from the use of multiple computing resources, with, for example, simulation codes running on highly parallel supercomputers and AI training and inference tasks on specialized accelerators. Here, we present our experiences deploying two AI-guided simulation workflows across such heterogeneous systems. A unique aspect of our approach is our use of cloud-hosted management services to manage challenging aspects of cross-resource authentication and authorization, function-as-a-service (FaaS) function invocation, and data transfer. We show that these methods can achieve performance parity with systems that rely on direct connection between resources. We achieve parity by integrating the FaaS system and data transfer capabilities with a system that passes data by reference among managers and workers, and a user-configurable steering algorithm to hide data transfer latencies. We anticipate that this ease of use can enable routine use of heterogeneous resources in computational science.
Colmena: Scalable Machine-Learning-Based Steering of Ensemble Simulations for High Performance Computing
Oct 06, 2021



Abstract:Scientific applications that involve simulation ensembles can be accelerated greatly by using experiment design methods to select the best simulations to perform. Methods that use machine learning (ML) to create proxy models of simulations show particular promise for guiding ensembles but are challenging to deploy because of the need to coordinate dynamic mixes of simulation and learning tasks. We present Colmena, an open-source Python framework that allows users to steer campaigns by providing just the implementations of individual tasks plus the logic used to choose which tasks to execute when. Colmena handles task dispatch, results collation, ML model invocation, and ML model (re)training, using Parsl to execute tasks on HPC systems. We describe the design of Colmena and illustrate its capabilities by applying it to electrolyte design, where it both scales to 65536 CPUs and accelerates the discovery rate for high-performance molecules by a factor of 100 over unguided searches.
KAISA: An Adaptive Second-order Optimizer Framework for Deep Neural Networks
Jul 04, 2021



Abstract:Kronecker-factored Approximate Curvature (K-FAC) has recently been shown to converge faster in deep neural network (DNN) training than stochastic gradient descent (SGD); however, K-FAC's larger memory footprint hinders its applicability to large models. We present KAISA, a K-FAC-enabled, Adaptable, Improved, and ScAlable second-order optimizer framework that adapts the memory footprint, communication, and computation given specific models and hardware to achieve maximized performance and enhanced scalability. We quantify the tradeoffs between memory and communication cost and evaluate KAISA on large models, including ResNet-50, Mask R-CNN, U-Net, and BERT, on up to 128 NVIDIA A100 GPUs. Compared to the original optimizers, KAISA converges 18.1-36.3% faster across applications with the same global batch size. Under a fixed memory budget, KAISA converges 32.5% and 41.6% faster in ResNet-50 and BERT-Large, respectively. KAISA can balance memory and communication to achieve scaling efficiency equal to or better than the baseline optimizers.
AI- and HPC-enabled Lead Generation for SARS-CoV-2: Models and Processes to Extract Druglike Molecules Contained in Natural Language Text
Jan 12, 2021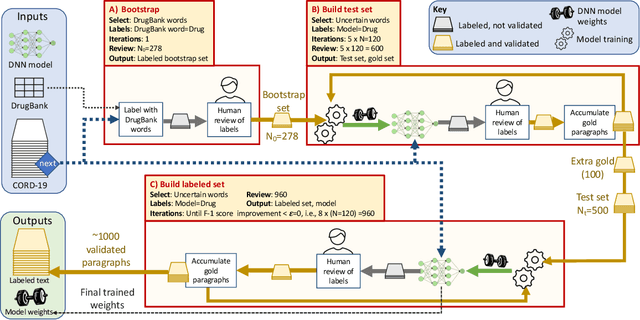
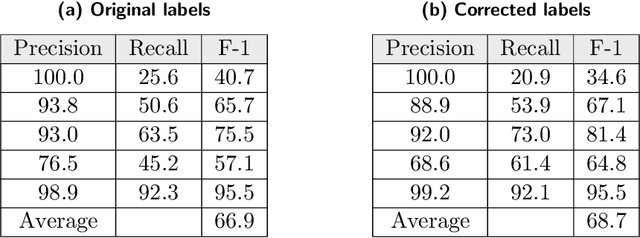
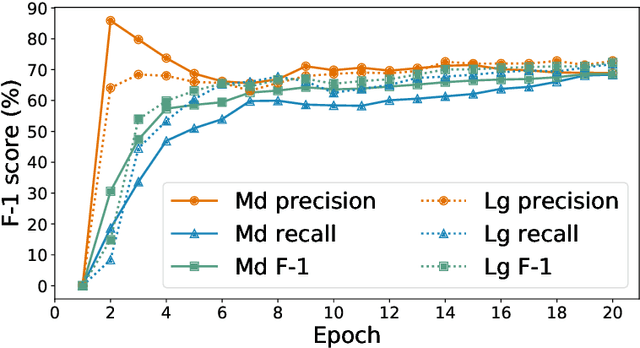
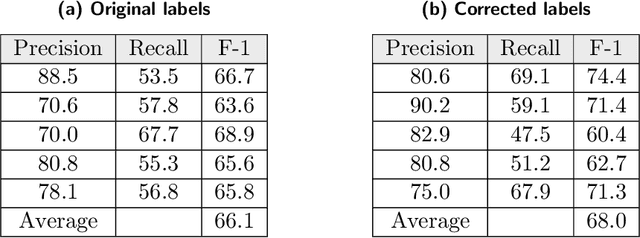
Abstract:Researchers worldwide are seeking to repurpose existing drugs or discover new drugs to counter the disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A promising source of candidates for such studies is molecules that have been reported in the scientific literature to be drug-like in the context of coronavirus research. We report here on a project that leverages both human and artificial intelligence to detect references to drug-like molecules in free text. We engage non-expert humans to create a corpus of labeled text, use this labeled corpus to train a named entity recognition model, and employ the trained model to extract 10912 drug-like molecules from the COVID-19 Open Research Dataset Challenge (CORD-19) corpus of 198875 papers. Performance analyses show that our automated extraction model can achieve performance on par with that of non-expert humans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge