Ian Foster
Topology-Aware Knowledge Propagation in Decentralized Learning
May 16, 2025Abstract:Decentralized learning enables collaborative training of models across naturally distributed data without centralized coordination or maintenance of a global model. Instead, devices are organized in arbitrary communication topologies, in which they can only communicate with neighboring devices. Each device maintains its own local model by training on its local data and integrating new knowledge via model aggregation with neighbors. Therefore, knowledge is propagated across the topology via successive aggregation rounds. We study, in particular, the propagation of out-of-distribution (OOD) knowledge. We find that popular decentralized learning algorithms struggle to propagate OOD knowledge effectively to all devices. Further, we find that both the location of OOD data within a topology, and the topology itself, significantly impact OOD knowledge propagation. We then propose topology-aware aggregation strategies to accelerate (OOD) knowledge propagation across devices. These strategies improve OOD data accuracy, compared to topology-unaware baselines, by 123% on average across models in a topology.
HiPerRAG: High-Performance Retrieval Augmented Generation for Scientific Insights
May 07, 2025Abstract:The volume of scientific literature is growing exponentially, leading to underutilized discoveries, duplicated efforts, and limited cross-disciplinary collaboration. Retrieval Augmented Generation (RAG) offers a way to assist scientists by improving the factuality of Large Language Models (LLMs) in processing this influx of information. However, scaling RAG to handle millions of articles introduces significant challenges, including the high computational costs associated with parsing documents and embedding scientific knowledge, as well as the algorithmic complexity of aligning these representations with the nuanced semantics of scientific content. To address these issues, we introduce HiPerRAG, a RAG workflow powered by high performance computing (HPC) to index and retrieve knowledge from more than 3.6 million scientific articles. At its core are Oreo, a high-throughput model for multimodal document parsing, and ColTrast, a query-aware encoder fine-tuning algorithm that enhances retrieval accuracy by using contrastive learning and late-interaction techniques. HiPerRAG delivers robust performance on existing scientific question answering benchmarks and two new benchmarks introduced in this work, achieving 90% accuracy on SciQ and 76% on PubMedQA-outperforming both domain-specific models like PubMedGPT and commercial LLMs such as GPT-4. Scaling to thousands of GPUs on the Polaris, Sunspot, and Frontier supercomputers, HiPerRAG delivers million document-scale RAG workflows for unifying scientific knowledge and fostering interdisciplinary innovation.
34 Examples of LLM Applications in Materials Science and Chemistry: Towards Automation, Assistants, Agents, and Accelerated Scientific Discovery
May 05, 2025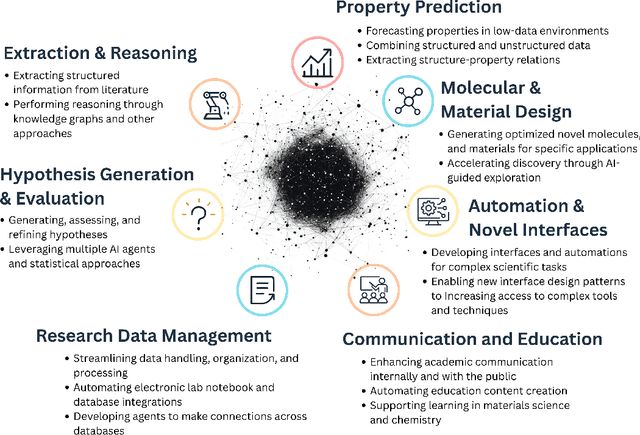
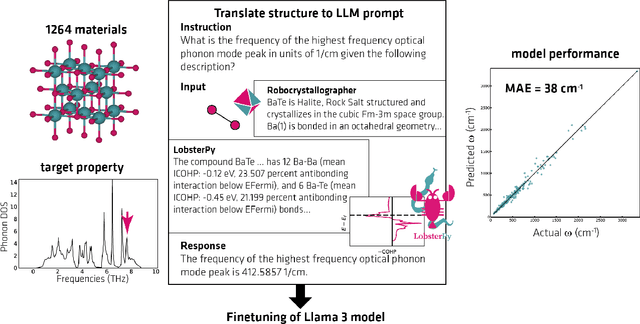
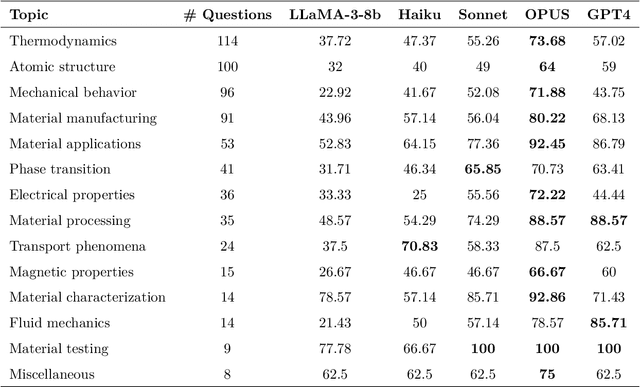
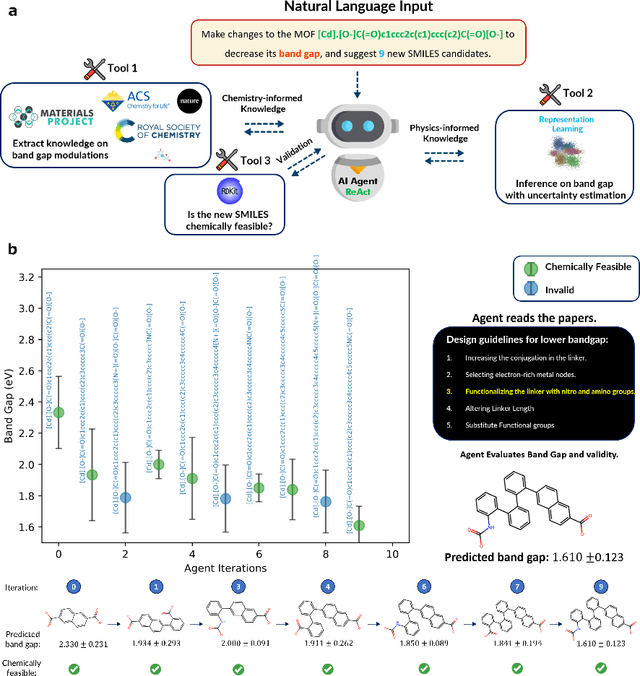
Abstract:Large Language Models (LLMs) are reshaping many aspects of materials science and chemistry research, enabling advances in molecular property prediction, materials design, scientific automation, knowledge extraction, and more. Recent developments demonstrate that the latest class of models are able to integrate structured and unstructured data, assist in hypothesis generation, and streamline research workflows. To explore the frontier of LLM capabilities across the research lifecycle, we review applications of LLMs through 34 total projects developed during the second annual Large Language Model Hackathon for Applications in Materials Science and Chemistry, a global hybrid event. These projects spanned seven key research areas: (1) molecular and material property prediction, (2) molecular and material design, (3) automation and novel interfaces, (4) scientific communication and education, (5) research data management and automation, (6) hypothesis generation and evaluation, and (7) knowledge extraction and reasoning from the scientific literature. Collectively, these applications illustrate how LLMs serve as versatile predictive models, platforms for rapid prototyping of domain-specific tools, and much more. In particular, improvements in both open source and proprietary LLM performance through the addition of reasoning, additional training data, and new techniques have expanded effectiveness, particularly in low-data environments and interdisciplinary research. As LLMs continue to improve, their integration into scientific workflows presents both new opportunities and new challenges, requiring ongoing exploration, continued refinement, and further research to address reliability, interpretability, and reproducibility.
AdaParse: An Adaptive Parallel PDF Parsing and Resource Scaling Engine
Apr 23, 2025Abstract:Language models for scientific tasks are trained on text from scientific publications, most distributed as PDFs that require parsing. PDF parsing approaches range from inexpensive heuristics (for simple documents) to computationally intensive ML-driven systems (for complex or degraded ones). The choice of the "best" parser for a particular document depends on its computational cost and the accuracy of its output. To address these issues, we introduce an Adaptive Parallel PDF Parsing and Resource Scaling Engine (AdaParse), a data-driven strategy for assigning an appropriate parser to each document. We enlist scientists to select preferred parser outputs and incorporate this information through direct preference optimization (DPO) into AdaParse, thereby aligning its selection process with human judgment. AdaParse then incorporates hardware requirements and predicted accuracy of each parser to orchestrate computational resources efficiently for large-scale parsing campaigns. We demonstrate that AdaParse, when compared to state-of-the-art parsers, improves throughput by $17\times$ while still achieving comparable accuracy (0.2 percent better) on a benchmark set of 1000 scientific documents. AdaParse's combination of high accuracy and parallel scalability makes it feasible to parse large-scale scientific document corpora to support the development of high-quality, trillion-token-scale text datasets. The implementation is available at https://github.com/7shoe/AdaParse/
Advances and Challenges in Foundation Agents: From Brain-Inspired Intelligence to Evolutionary, Collaborative, and Safe Systems
Mar 31, 2025Abstract:The advent of large language models (LLMs) has catalyzed a transformative shift in artificial intelligence, paving the way for advanced intelligent agents capable of sophisticated reasoning, robust perception, and versatile action across diverse domains. As these agents increasingly drive AI research and practical applications, their design, evaluation, and continuous improvement present intricate, multifaceted challenges. This survey provides a comprehensive overview, framing intelligent agents within a modular, brain-inspired architecture that integrates principles from cognitive science, neuroscience, and computational research. We structure our exploration into four interconnected parts. First, we delve into the modular foundation of intelligent agents, systematically mapping their cognitive, perceptual, and operational modules onto analogous human brain functionalities, and elucidating core components such as memory, world modeling, reward processing, and emotion-like systems. Second, we discuss self-enhancement and adaptive evolution mechanisms, exploring how agents autonomously refine their capabilities, adapt to dynamic environments, and achieve continual learning through automated optimization paradigms, including emerging AutoML and LLM-driven optimization strategies. Third, we examine collaborative and evolutionary multi-agent systems, investigating the collective intelligence emerging from agent interactions, cooperation, and societal structures, highlighting parallels to human social dynamics. Finally, we address the critical imperative of building safe, secure, and beneficial AI systems, emphasizing intrinsic and extrinsic security threats, ethical alignment, robustness, and practical mitigation strategies necessary for trustworthy real-world deployment.
EAIRA: Establishing a Methodology for Evaluating AI Models as Scientific Research Assistants
Feb 27, 2025Abstract:Recent advancements have positioned AI, and particularly Large Language Models (LLMs), as transformative tools for scientific research, capable of addressing complex tasks that require reasoning, problem-solving, and decision-making. Their exceptional capabilities suggest their potential as scientific research assistants but also highlight the need for holistic, rigorous, and domain-specific evaluation to assess effectiveness in real-world scientific applications. This paper describes a multifaceted methodology for Evaluating AI models as scientific Research Assistants (EAIRA) developed at Argonne National Laboratory. This methodology incorporates four primary classes of evaluations. 1) Multiple Choice Questions to assess factual recall; 2) Open Response to evaluate advanced reasoning and problem-solving skills; 3) Lab-Style Experiments involving detailed analysis of capabilities as research assistants in controlled environments; and 4) Field-Style Experiments to capture researcher-LLM interactions at scale in a wide range of scientific domains and applications. These complementary methods enable a comprehensive analysis of LLM strengths and weaknesses with respect to their scientific knowledge, reasoning abilities, and adaptability. Recognizing the rapid pace of LLM advancements, we designed the methodology to evolve and adapt so as to ensure its continued relevance and applicability. This paper describes the methodology state at the end of February 2025. Although developed within a subset of scientific domains, the methodology is designed to be generalizable to a wide range of scientific domains.
Connecting Large Language Model Agent to High Performance Computing Resource
Feb 17, 2025Abstract:The Large Language Model agent workflow enables the LLM to invoke tool functions to increase the performance on specific scientific domain questions. To tackle large scale of scientific research, it requires access to computing resource and parallel computing setup. In this work, we implemented Parsl to the LangChain/LangGraph tool call setup, to bridge the gap between the LLM agent to the computing resource. Two tool call implementations were set up and tested on both local workstation and HPC environment on Polaris/ALCF. The first implementation with Parsl-enabled LangChain tool node queues the tool functions concurrently to the Parsl workers for parallel execution. The second configuration is implemented by converting the tool functions into Parsl ensemble functions, and is more suitable for large task on super computer environment. The LLM agent workflow was prompted to run molecular dynamics simulations, with different protein structure and simulation conditions. These results showed the LLM agent tools were managed and executed concurrently by Parsl on the available computing resource.
DrugImproverGPT: A Large Language Model for Drug Optimization with Fine-Tuning via Structured Policy Optimization
Feb 11, 2025Abstract:Finetuning a Large Language Model (LLM) is crucial for generating results towards specific objectives. This research delves into the realm of drug optimization and introduce a novel reinforcement learning algorithm to finetune a drug optimization LLM-based generative model, enhancing the original drug across target objectives, while retains the beneficial chemical properties of the original drug. This work is comprised of two primary components: (1) DrugImprover: A framework tailored for improving robustness and efficiency in drug optimization. It includes a LLM designed for drug optimization and a novel Structured Policy Optimization (SPO) algorithm, which is theoretically grounded. This algorithm offers a unique perspective for fine-tuning the LLM-based generative model by aligning the improvement of the generated molecule with the input molecule under desired objectives. (2) A dataset of 1 million compounds, each with OEDOCK docking scores on 5 human proteins associated with cancer cells and 24 binding sites from SARS-CoV-2 virus. We conduct a comprehensive evaluation of SPO and demonstrate its effectiveness in improving the original drug across target properties. Our code and dataset will be publicly available at: https://github.com/xuefeng-cs/DrugImproverGPT.
MOFA: Discovering Materials for Carbon Capture with a GenAI- and Simulation-Based Workflow
Jan 18, 2025Abstract:We present MOFA, an open-source generative AI (GenAI) plus simulation workflow for high-throughput generation of metal-organic frameworks (MOFs) on large-scale high-performance computing (HPC) systems. MOFA addresses key challenges in integrating GPU-accelerated computing for GPU-intensive GenAI tasks, including distributed training and inference, alongside CPU- and GPU-optimized tasks for screening and filtering AI-generated MOFs using molecular dynamics, density functional theory, and Monte Carlo simulations. These heterogeneous tasks are unified within an online learning framework that optimizes the utilization of available CPU and GPU resources across HPC systems. Performance metrics from a 450-node (14,400 AMD Zen 3 CPUs + 1800 NVIDIA A100 GPUs) supercomputer run demonstrate that MOFA achieves high-throughput generation of novel MOF structures, with CO$_2$ adsorption capacities ranking among the top 10 in the hypothetical MOF (hMOF) dataset. Furthermore, the production of high-quality MOFs exhibits a linear relationship with the number of nodes utilized. The modular architecture of MOFA will facilitate its integration into other scientific applications that dynamically combine GenAI with large-scale simulations.
Reflections from the 2024 Large Language Model (LLM) Hackathon for Applications in Materials Science and Chemistry
Nov 20, 2024
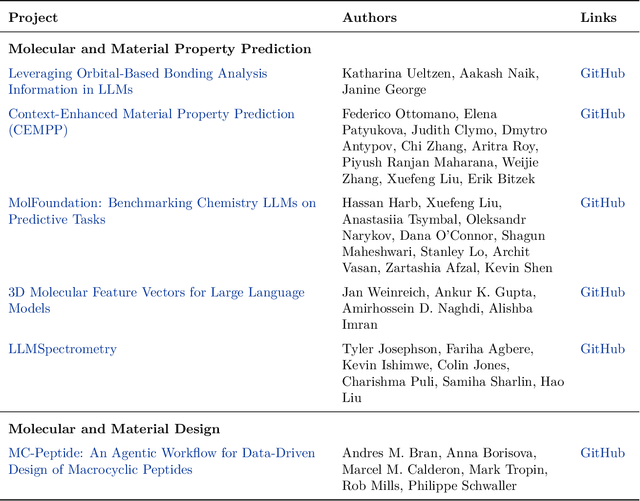
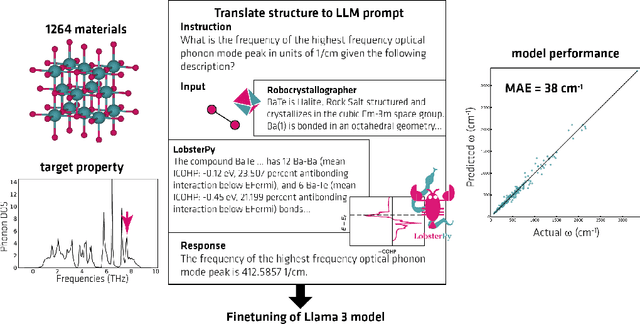
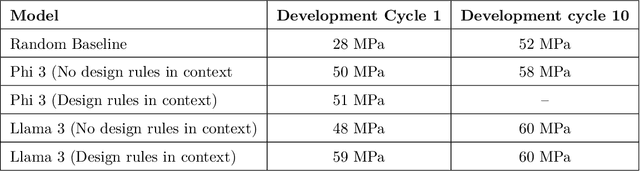
Abstract:Here, we present the outcomes from the second Large Language Model (LLM) Hackathon for Applications in Materials Science and Chemistry, which engaged participants across global hybrid locations, resulting in 34 team submissions. The submissions spanned seven key application areas and demonstrated the diverse utility of LLMs for applications in (1) molecular and material property prediction; (2) molecular and material design; (3) automation and novel interfaces; (4) scientific communication and education; (5) research data management and automation; (6) hypothesis generation and evaluation; and (7) knowledge extraction and reasoning from scientific literature. Each team submission is presented in a summary table with links to the code and as brief papers in the appendix. Beyond team results, we discuss the hackathon event and its hybrid format, which included physical hubs in Toronto, Montreal, San Francisco, Berlin, Lausanne, and Tokyo, alongside a global online hub to enable local and virtual collaboration. Overall, the event highlighted significant improvements in LLM capabilities since the previous year's hackathon, suggesting continued expansion of LLMs for applications in materials science and chemistry research. These outcomes demonstrate the dual utility of LLMs as both multipurpose models for diverse machine learning tasks and platforms for rapid prototyping custom applications in scientific research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge