Yadu Babuji
Scalable Agentic Reasoning for Designing Biologics Targeting Intrinsically Disordered Proteins
Dec 17, 2025Abstract:Intrinsically disordered proteins (IDPs) represent crucial therapeutic targets due to their significant role in disease -- approximately 80\% of cancer-related proteins contain long disordered regions -- but their lack of stable secondary/tertiary structures makes them "undruggable". While recent computational advances, such as diffusion models, can design high-affinity IDP binders, translating these to practical drug discovery requires autonomous systems capable of reasoning across complex conformational ensembles and orchestrating diverse computational tools at scale.To address this challenge, we designed and implemented StructBioReasoner, a scalable multi-agent system for designing biologics that can be used to target IDPs. StructBioReasoner employs a novel tournament-based reasoning framework where specialized agents compete to generate and refine therapeutic hypotheses, naturally distributing computational load for efficient exploration of the vast design space. Agents integrate domain knowledge with access to literature synthesis, AI-structure prediction, molecular simulations, and stability analysis, coordinating their execution on HPC infrastructure via an extensible federated agentic middleware, Academy. We benchmark StructBioReasoner across Der f 21 and NMNAT-2 and demonstrate that over 50\% of 787 designed and validated candidates for Der f 21 outperformed the human-designed reference binders from literature, in terms of improved binding free energy. For the more challenging NMNAT-2 protein, we identified three binding modes from 97,066 binders, including the well-studied NMNAT2:p53 interface. Thus, StructBioReasoner lays the groundwork for agentic reasoning systems for IDP therapeutic discovery on Exascale platforms.
AdaParse: An Adaptive Parallel PDF Parsing and Resource Scaling Engine
Apr 23, 2025Abstract:Language models for scientific tasks are trained on text from scientific publications, most distributed as PDFs that require parsing. PDF parsing approaches range from inexpensive heuristics (for simple documents) to computationally intensive ML-driven systems (for complex or degraded ones). The choice of the "best" parser for a particular document depends on its computational cost and the accuracy of its output. To address these issues, we introduce an Adaptive Parallel PDF Parsing and Resource Scaling Engine (AdaParse), a data-driven strategy for assigning an appropriate parser to each document. We enlist scientists to select preferred parser outputs and incorporate this information through direct preference optimization (DPO) into AdaParse, thereby aligning its selection process with human judgment. AdaParse then incorporates hardware requirements and predicted accuracy of each parser to orchestrate computational resources efficiently for large-scale parsing campaigns. We demonstrate that AdaParse, when compared to state-of-the-art parsers, improves throughput by $17\times$ while still achieving comparable accuracy (0.2 percent better) on a benchmark set of 1000 scientific documents. AdaParse's combination of high accuracy and parallel scalability makes it feasible to parse large-scale scientific document corpora to support the development of high-quality, trillion-token-scale text datasets. The implementation is available at https://github.com/7shoe/AdaParse/
LSHBloom: Memory-efficient, Extreme-scale Document Deduplication
Nov 06, 2024Abstract:Deduplication is a major focus for assembling and curating training datasets for large language models (LLM) -- detecting and eliminating additional instances of the same content -- in large collections of technical documents. Unrestrained, duplicates in the training dataset increase training costs and lead to undesirable properties such as memorization in trained models or cheating on evaluation. Contemporary approaches to document-level deduplication are often extremely expensive in both runtime and memory. We propose LSHBloom, an extension to MinhashLSH, which replaces the expensive LSHIndex with lightweight Bloom filters. LSHBloom demonstrates the same deduplication performance as MinhashLSH with only a marginal increase in false positives (as low as 1e-5 in our experiments); demonstrates competitive runtime (270\% faster than MinhashLSH on peS2o); and, crucially, uses just 0.6\% of the disk space required by MinhashLSH to deduplicate peS2o. We demonstrate that this space advantage scales with increased dataset size -- at the extreme scale of several billion documents, LSHBloom promises a 250\% speedup and a 54$\times$ space advantage over traditional MinHashLSH scaling deduplication of text datasets to many billions of documents.
Flight: A FaaS-Based Framework for Complex and Hierarchical Federated Learning
Sep 24, 2024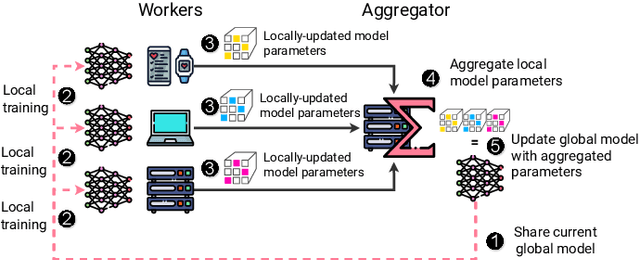
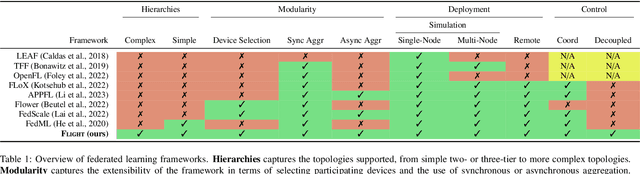
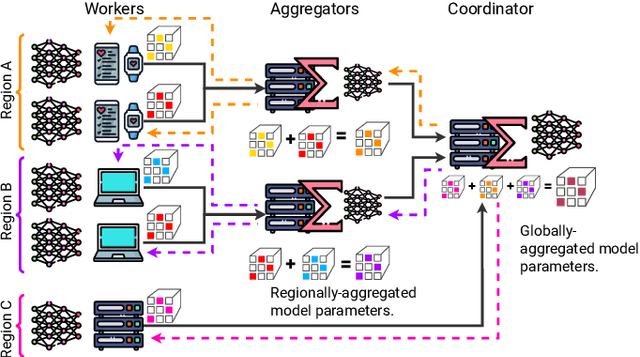
Abstract:Federated Learning (FL) is a decentralized machine learning paradigm where models are trained on distributed devices and are aggregated at a central server. Existing FL frameworks assume simple two-tier network topologies where end devices are directly connected to the aggregation server. While this is a practical mental model, it does not exploit the inherent topology of real-world distributed systems like the Internet-of-Things. We present Flight, a novel FL framework that supports complex hierarchical multi-tier topologies, asynchronous aggregation, and decouples the control plane from the data plane. We compare the performance of Flight against Flower, a state-of-the-art FL framework. Our results show that Flight scales beyond Flower, supporting up to 2048 simultaneous devices, and reduces FL makespan across several models. Finally, we show that Flight's hierarchical FL model can reduce communication overheads by more than 60%.
Employing Artificial Intelligence to Steer Exascale Workflows with Colmena
Aug 26, 2024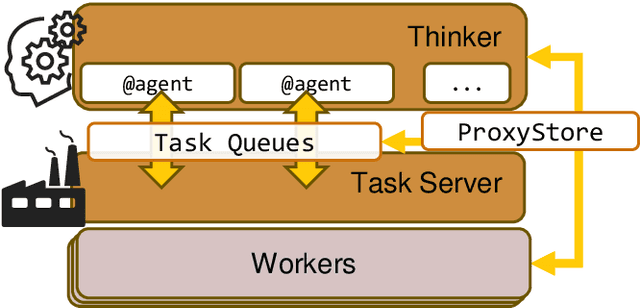
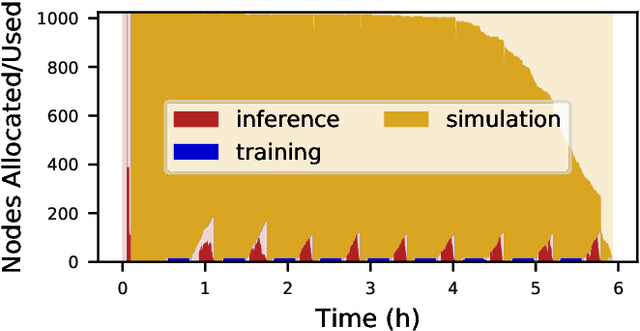
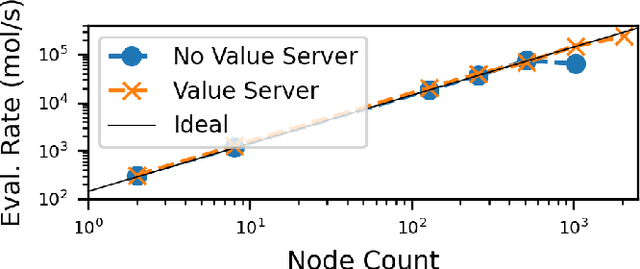
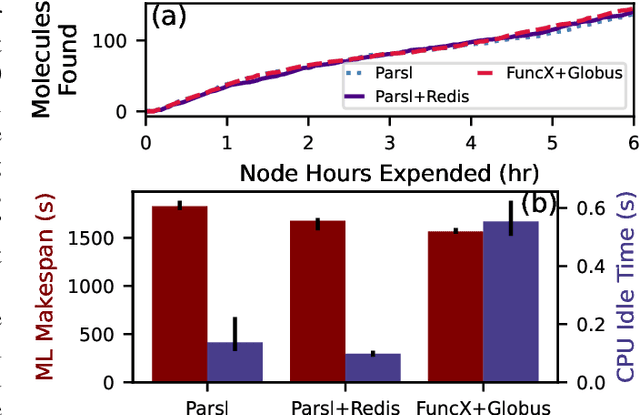
Abstract:Computational workflows are a common class of application on supercomputers, yet the loosely coupled and heterogeneous nature of workflows often fails to take full advantage of their capabilities. We created Colmena to leverage the massive parallelism of a supercomputer by using Artificial Intelligence (AI) to learn from and adapt a workflow as it executes. Colmena allows scientists to define how their application should respond to events (e.g., task completion) as a series of cooperative agents. In this paper, we describe the design of Colmena, the challenges we overcame while deploying applications on exascale systems, and the science workflows we have enhanced through interweaving AI. The scaling challenges we discuss include developing steering strategies that maximize node utilization, introducing data fabrics that reduce communication overhead of data-intensive tasks, and implementing workflow tasks that cache costly operations between invocations. These innovations coupled with a variety of application patterns accessible through our agent-based steering model have enabled science advances in chemistry, biophysics, and materials science using different types of AI. Our vision is that Colmena will spur creative solutions that harness AI across many domains of scientific computing.
Cloud Services Enable Efficient AI-Guided Simulation Workflows across Heterogeneous Resources
Mar 15, 2023Abstract:Applications that fuse machine learning and simulation can benefit from the use of multiple computing resources, with, for example, simulation codes running on highly parallel supercomputers and AI training and inference tasks on specialized accelerators. Here, we present our experiences deploying two AI-guided simulation workflows across such heterogeneous systems. A unique aspect of our approach is our use of cloud-hosted management services to manage challenging aspects of cross-resource authentication and authorization, function-as-a-service (FaaS) function invocation, and data transfer. We show that these methods can achieve performance parity with systems that rely on direct connection between resources. We achieve parity by integrating the FaaS system and data transfer capabilities with a system that passes data by reference among managers and workers, and a user-configurable steering algorithm to hide data transfer latencies. We anticipate that this ease of use can enable routine use of heterogeneous resources in computational science.
Colmena: Scalable Machine-Learning-Based Steering of Ensemble Simulations for High Performance Computing
Oct 06, 2021



Abstract:Scientific applications that involve simulation ensembles can be accelerated greatly by using experiment design methods to select the best simulations to perform. Methods that use machine learning (ML) to create proxy models of simulations show particular promise for guiding ensembles but are challenging to deploy because of the need to coordinate dynamic mixes of simulation and learning tasks. We present Colmena, an open-source Python framework that allows users to steer campaigns by providing just the implementations of individual tasks plus the logic used to choose which tasks to execute when. Colmena handles task dispatch, results collation, ML model invocation, and ML model (re)training, using Parsl to execute tasks on HPC systems. We describe the design of Colmena and illustrate its capabilities by applying it to electrolyte design, where it both scales to 65536 CPUs and accelerates the discovery rate for high-performance molecules by a factor of 100 over unguided searches.
Protein-Ligand Docking Surrogate Models: A SARS-CoV-2 Benchmark for Deep Learning Accelerated Virtual Screening
Jun 30, 2021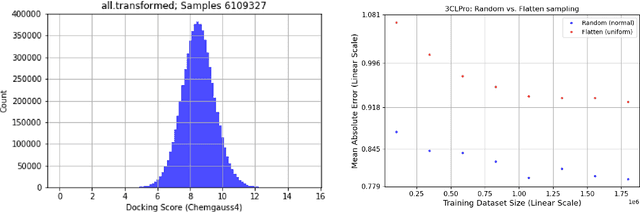
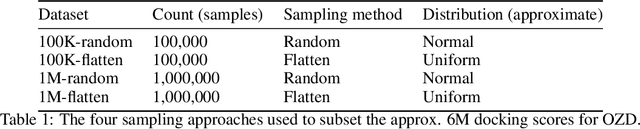
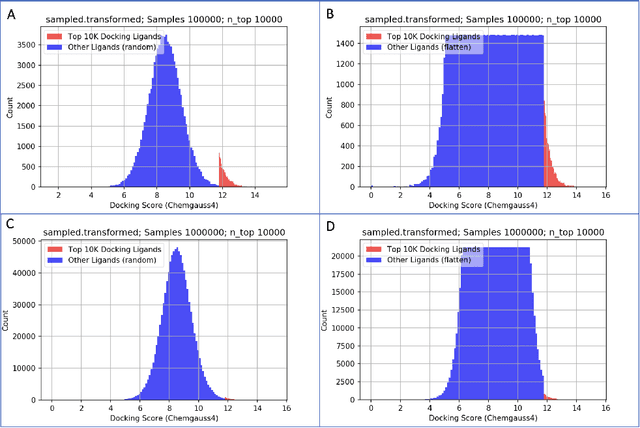
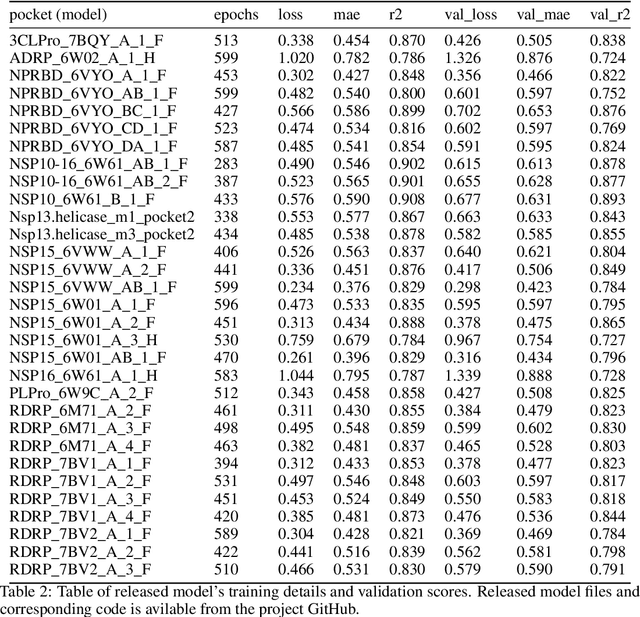
Abstract:We propose a benchmark to study surrogate model accuracy for protein-ligand docking. We share a dataset consisting of 200 million 3D complex structures and 2D structure scores across a consistent set of 13 million "in-stock" molecules over 15 receptors, or binding sites, across the SARS-CoV-2 proteome. Our work shows surrogate docking models have six orders of magnitude more throughput than standard docking protocols on the same supercomputer node types. We demonstrate the power of high-speed surrogate models by running each target against 1 billion molecules in under a day (50k predictions per GPU seconds). We showcase a workflow for docking utilizing surrogate ML models as a pre-filter. Our workflow is ten times faster at screening a library of compounds than the standard technique, with an error rate less than 0.01\% of detecting the underlying best scoring 0.1\% of compounds. Our analysis of the speedup explains that to screen more molecules under a docking paradigm, another order of magnitude speedup must come from model accuracy rather than computing speed (which, if increased, will not anymore alter our throughput to screen molecules). We believe this is strong evidence for the community to begin focusing on improving the accuracy of surrogate models to improve the ability to screen massive compound libraries 100x or even 1000x faster than current techniques.
Targeting SARS-CoV-2 with AI- and HPC-enabled Lead Generation: A First Data Release
May 28, 2020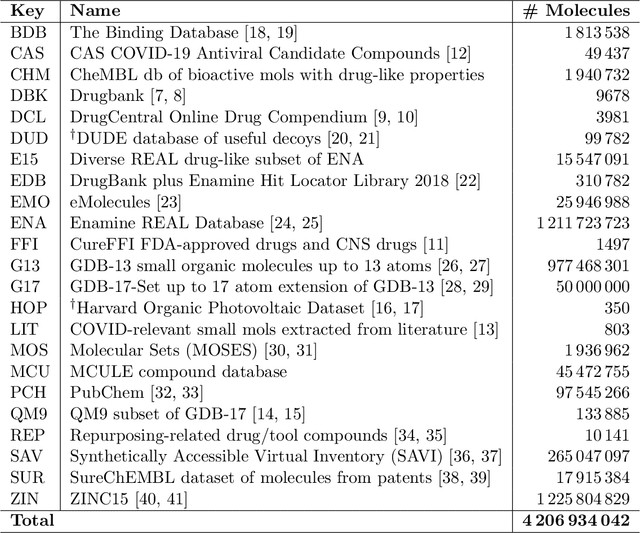
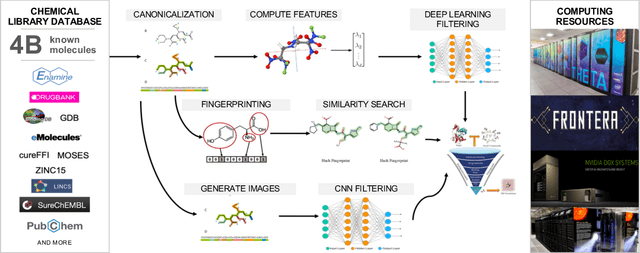
Abstract:Researchers across the globe are seeking to rapidly repurpose existing drugs or discover new drugs to counter the the novel coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). One promising approach is to train machine learning (ML) and artificial intelligence (AI) tools to screen large numbers of small molecules. As a contribution to that effort, we are aggregating numerous small molecules from a variety of sources, using high-performance computing (HPC) to computer diverse properties of those molecules, using the computed properties to train ML/AI models, and then using the resulting models for screening. In this first data release, we make available 23 datasets collected from community sources representing over 4.2 B molecules enriched with pre-computed: 1) molecular fingerprints to aid similarity searches, 2) 2D images of molecules to enable exploration and application of image-based deep learning methods, and 3) 2D and 3D molecular descriptors to speed development of machine learning models. This data release encompasses structural information on the 4.2 B molecules and 60 TB of pre-computed data. Future releases will expand the data to include more detailed molecular simulations, computed models, and other products.
DLHub: Model and Data Serving for Science
Nov 27, 2018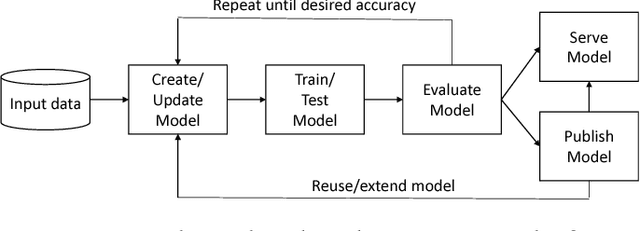
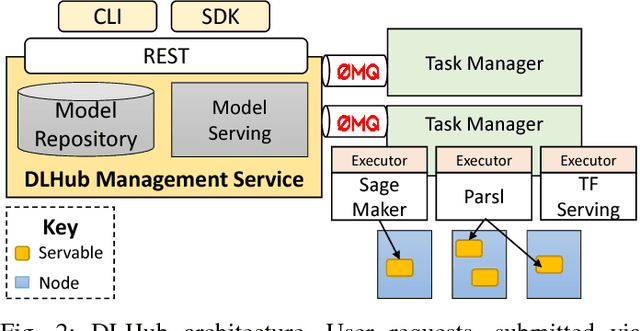
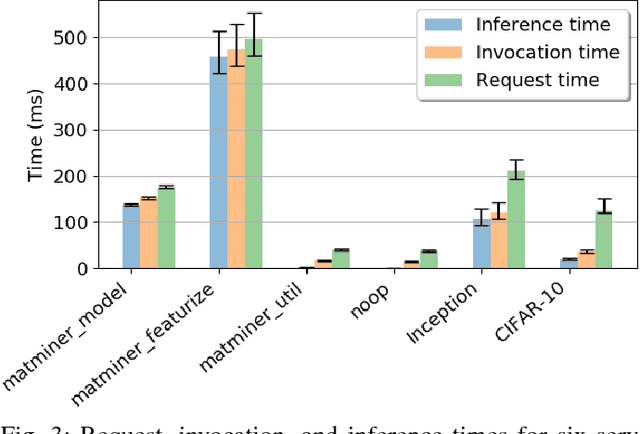
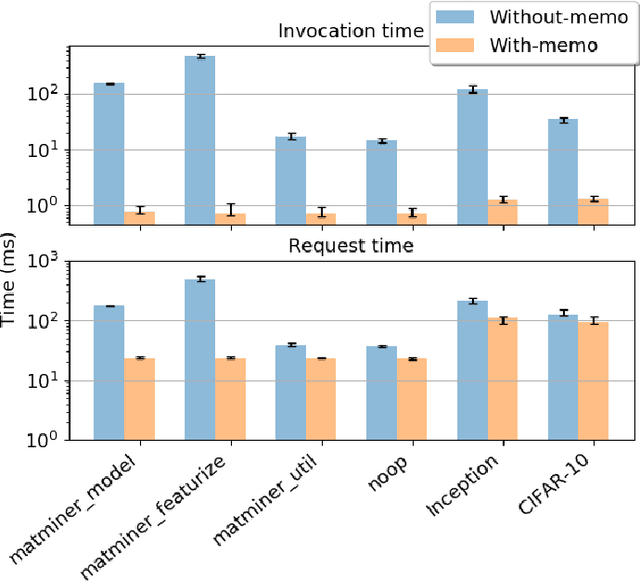
Abstract:While the Machine Learning (ML) landscape is evolving rapidly, there has been a relative lag in the development of the "learning systems" needed to enable broad adoption. Furthermore, few such systems are designed to support the specialized requirements of scientific ML. Here we present the Data and Learning Hub for science (DLHub), a multi-tenant system that provides both model repository and serving capabilities with a focus on science applications. DLHub addresses two significant shortcomings in current systems. First, its selfservice model repository allows users to share, publish, verify, reproduce, and reuse models, and addresses concerns related to model reproducibility by packaging and distributing models and all constituent components. Second, it implements scalable and low-latency serving capabilities that can leverage parallel and distributed computing resources to democratize access to published models through a simple web interface. Unlike other model serving frameworks, DLHub can store and serve any Python 3-compatible model or processing function, plus multiple-function pipelines. We show that relative to other model serving systems including TensorFlow Serving, SageMaker, and Clipper, DLHub provides greater capabilities, comparable performance without memoization and batching, and significantly better performance when the latter two techniques can be employed. We also describe early uses of DLHub for scientific applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge