Zhuozhao Li
Unleashing the Potential of Differential Evolution through Individual-Level Strategy Diversity
Feb 01, 2026Abstract:Since Differential Evolution (DE) is sensitive to strategy choice, most existing variants pursue performance through adaptive mechanisms or intricate designs. While these approaches focus on adjusting strategies over time, the structural benefits that static strategy diversity may bring remain largely unexplored. To bridge this gap, we study the impact of individual-level strategy diversity on DE's search dynamics and performance, and introduce iStratDE (DE with individual-level strategies), a minimalist variant that assigns mutation and crossover strategies independently to each individual at initialization and keeps them fixed throughout the evolutionary process. By injecting diversity at the individual level without adaptation or feedback, iStratDE cultivates persistent behavioral heterogeneity that is especially effective with large populations. Moreover, its communication-free construction possesses intrinsic concurrency, thereby enabling efficient parallel execution and straightforward scaling for GPU computing. We further provide a convergence analysis of iStratDE under standard reachability assumptions, which establishes the almost-sure convergence of the best-so-far fitness. Extensive experiments on the CEC2022 benchmark suite and robotic control tasks demonstrate that iStratDE matches or surpasses established adaptive DE variants. These results highlight individual-level strategy assignment as a straightforward yet effective mechanism for enhancing DE's performance. The source code of iStratDE is publicly accessible at: https://github.com/EMI-Group/istratde.
Targeting SARS-CoV-2 with AI- and HPC-enabled Lead Generation: A First Data Release
May 28, 2020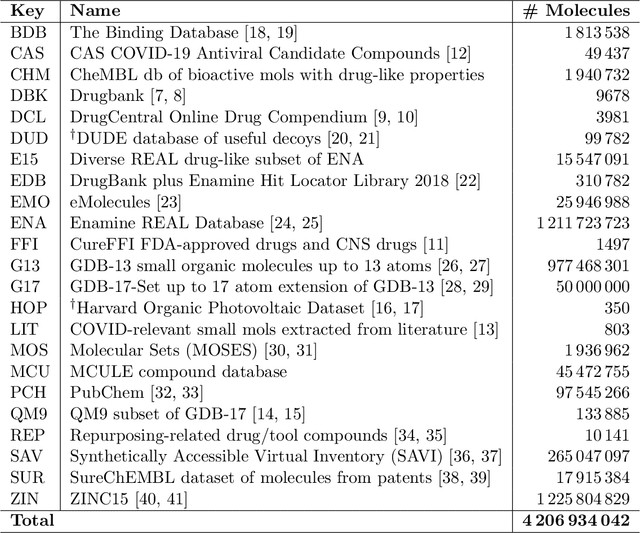
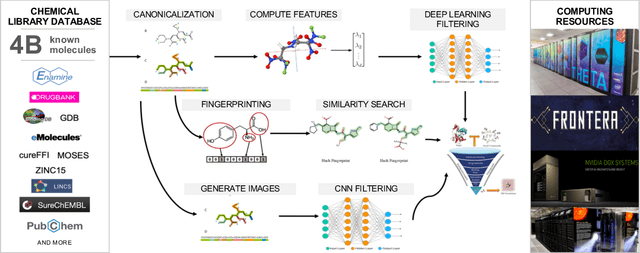
Abstract:Researchers across the globe are seeking to rapidly repurpose existing drugs or discover new drugs to counter the the novel coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). One promising approach is to train machine learning (ML) and artificial intelligence (AI) tools to screen large numbers of small molecules. As a contribution to that effort, we are aggregating numerous small molecules from a variety of sources, using high-performance computing (HPC) to computer diverse properties of those molecules, using the computed properties to train ML/AI models, and then using the resulting models for screening. In this first data release, we make available 23 datasets collected from community sources representing over 4.2 B molecules enriched with pre-computed: 1) molecular fingerprints to aid similarity searches, 2) 2D images of molecules to enable exploration and application of image-based deep learning methods, and 3) 2D and 3D molecular descriptors to speed development of machine learning models. This data release encompasses structural information on the 4.2 B molecules and 60 TB of pre-computed data. Future releases will expand the data to include more detailed molecular simulations, computed models, and other products.
DLHub: Model and Data Serving for Science
Nov 27, 2018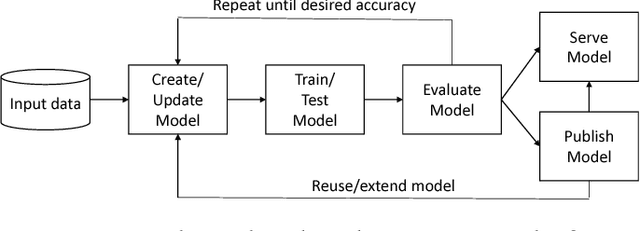
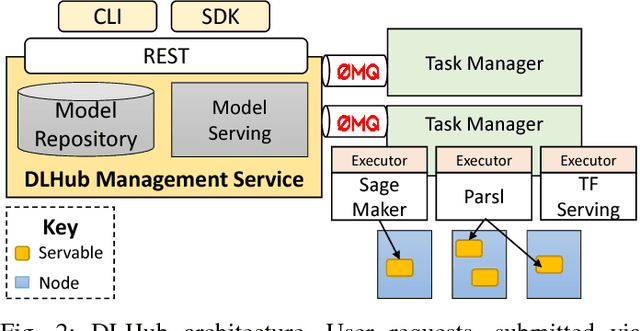
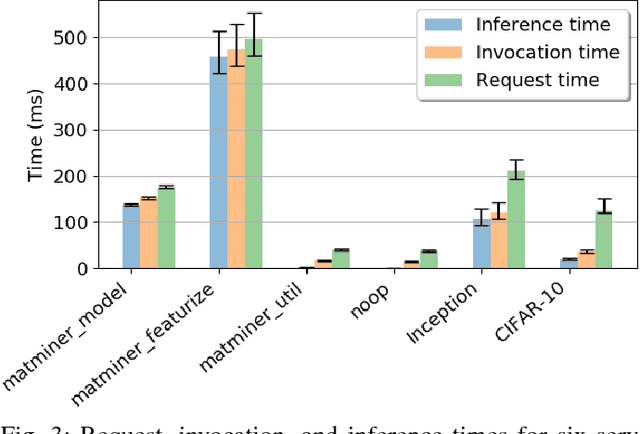
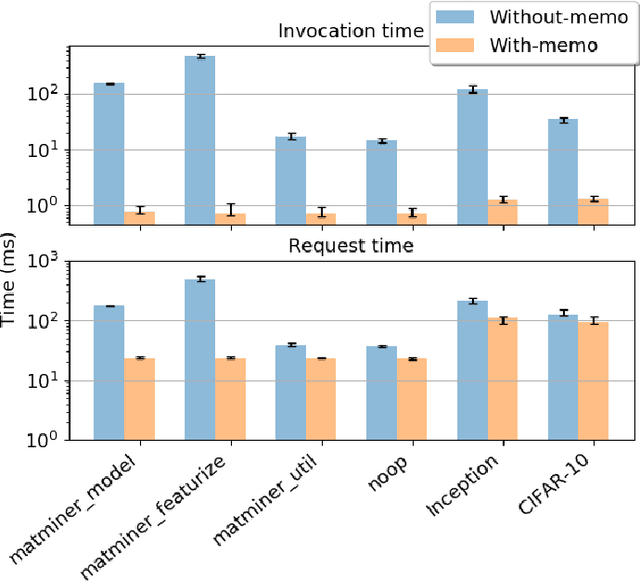
Abstract:While the Machine Learning (ML) landscape is evolving rapidly, there has been a relative lag in the development of the "learning systems" needed to enable broad adoption. Furthermore, few such systems are designed to support the specialized requirements of scientific ML. Here we present the Data and Learning Hub for science (DLHub), a multi-tenant system that provides both model repository and serving capabilities with a focus on science applications. DLHub addresses two significant shortcomings in current systems. First, its selfservice model repository allows users to share, publish, verify, reproduce, and reuse models, and addresses concerns related to model reproducibility by packaging and distributing models and all constituent components. Second, it implements scalable and low-latency serving capabilities that can leverage parallel and distributed computing resources to democratize access to published models through a simple web interface. Unlike other model serving frameworks, DLHub can store and serve any Python 3-compatible model or processing function, plus multiple-function pipelines. We show that relative to other model serving systems including TensorFlow Serving, SageMaker, and Clipper, DLHub provides greater capabilities, comparable performance without memoization and batching, and significantly better performance when the latter two techniques can be employed. We also describe early uses of DLHub for scientific applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge