Adib Bazgir
Autonomous Multi-Agent AI for High-Throughput Polymer Informatics: From Property Prediction to Generative Design Across Synthetic and Bio-Polymers
Jan 25, 2026Abstract:We present an integrated multiagent AI ecosystem for polymer discovery that unifies high-throughput materials workflows, artificial intelligence, and computational modeling within a single Polymer Research Lifecycle (PRL) pipeline. The system orchestrates specialized agents powered by state-of-the-art large language models (DeepSeek-V2 and DeepSeek-Coder) to retrieve and reason over scientific resources, invoke external tools, execute domain-specific code, and perform metacognitive self-assessment for robust end-to-end task execution. We demonstrate three practical capabilities: a high-fidelity polymer property prediction and generative design pipeline, a fully automated multimodal workflow for biopolymer structure characterization, and a metacognitive agent framework that can monitor performance and improve execution strategies over time. On a held-out test set of 1,251 polymers, our PolyGNN agent achieves strong predictive accuracy, reaching R2 = 0.89 for glass-transition temperature (Tg ), R2 = 0.82 for tensile strength, R2 = 0.75 for elongation, and R2 = 0.91 for density. The framework also provides uncertainty estimates via multiagent consensus and scales with linear complexity to at least 10,000 polymers, enabling high-throughput screening at low computational cost. For a representative workload, the system completes inference in 16.3 s using about 2 GB of memory and 0.1 GPU hours, at an estimated cost of about $0.08. On a dedicated Tg benchmark, our approach attains R2 = 0.78, outperforming strong baselines including single-LLM prediction (R2 = 0.67), group-contribution methods (R2 = 0.71), and ChemCrow (R2 = 0.66). We further demonstrate metacognitive control in a polystyrene case study, where the system not only produces domain-level scientific outputs but continually monitors and optimizes its own behavior through tactical, strategic, and meta-strategic self-assessment.
Quantum Error Mitigation with Attention Graph Transformers for Burgers Equation Solvers on NISQ Hardware
Dec 29, 2025Abstract:We present a hybrid quantum-classical framework augmented with learned error mitigation for solving the viscous Burgers equation on noisy intermediate-scale quantum (NISQ) hardware. Using the Cole-Hopf transformation, the nonlinear Burgers equation is mapped to a diffusion equation, discretized on uniform grids, and encoded into a quantum state whose time evolution is approximated via Trotterized nearest-neighbor circuits implemented in Qiskit. Quantum simulations are executed on noisy Aer backends and IBM superconducting quantum devices and are benchmarked against high-accuracy classical solutions obtained using a Krylov-based solver applied to the corresponding discretized Hamiltonian. From measured quantum amplitudes, we reconstruct the velocity field and evaluate physical and numerical diagnostics, including the L2 error, shock location, and dissipation rate, both with and without zero-noise extrapolation (ZNE). To enable data-driven error mitigation, we construct a large parametric dataset by sweeping viscosity, time step, grid resolution, and boundary conditions, producing matched tuples of noisy, ZNE-corrected, hardware, and classical solutions together with detailed circuit metadata. Leveraging this dataset, we train an attention-based graph neural network that incorporates circuit structure, light-cone information, global circuit parameters, and noisy quantum outputs to predict error-mitigated solutions. Across a wide range of parameters, the learned model consistently reduces the discrepancy between quantum and classical solutions beyond what is achieved by ZNE alone. We discuss extensions of this approach to higher-dimensional Burgers systems and more general quantum partial differential equation solvers, highlighting learned error mitigation as a promising complement to physics-based noise reduction techniques on NISQ devices.
Mic-hackathon 2024: Hackathon on Machine Learning for Electron and Scanning Probe Microscopy
Jun 10, 2025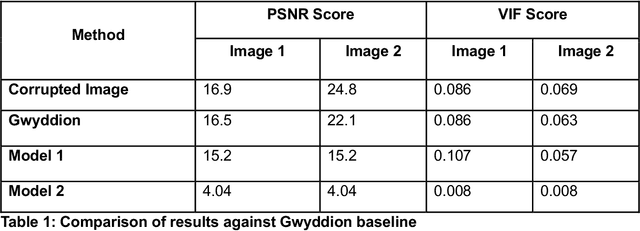
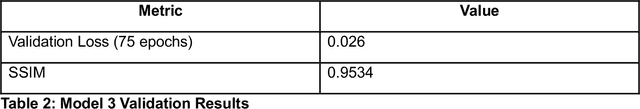
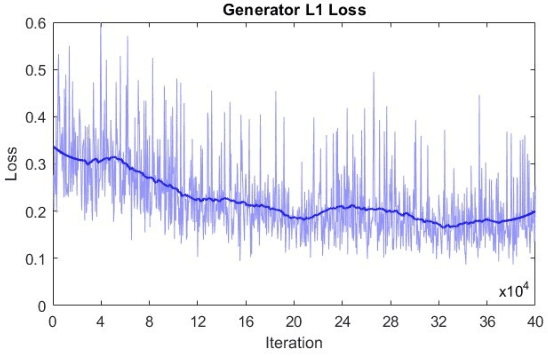
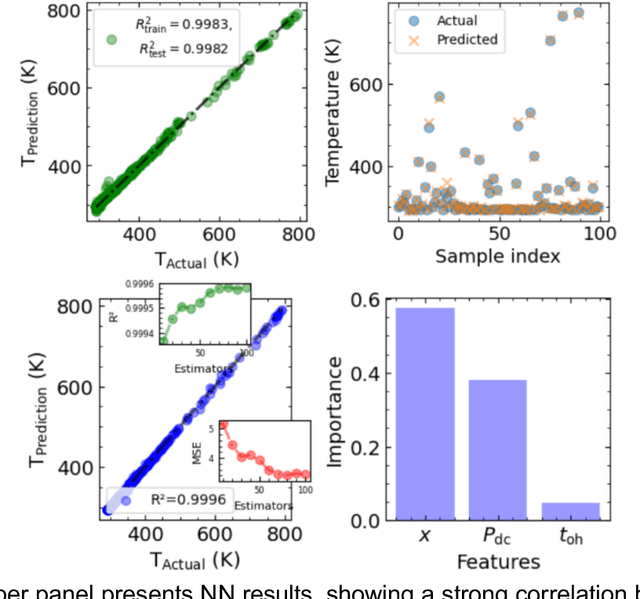
Abstract:Microscopy is a primary source of information on materials structure and functionality at nanometer and atomic scales. The data generated is often well-structured, enriched with metadata and sample histories, though not always consistent in detail or format. The adoption of Data Management Plans (DMPs) by major funding agencies promotes preservation and access. However, deriving insights remains difficult due to the lack of standardized code ecosystems, benchmarks, and integration strategies. As a result, data usage is inefficient and analysis time is extensive. In addition to post-acquisition analysis, new APIs from major microscope manufacturers enable real-time, ML-based analytics for automated decision-making and ML-agent-controlled microscope operation. Yet, a gap remains between the ML and microscopy communities, limiting the impact of these methods on physics, materials discovery, and optimization. Hackathons help bridge this divide by fostering collaboration between ML researchers and microscopy experts. They encourage the development of novel solutions that apply ML to microscopy, while preparing a future workforce for instrumentation, materials science, and applied ML. This hackathon produced benchmark datasets and digital twins of microscopes to support community growth and standardized workflows. All related code is available at GitHub: https://github.com/KalininGroup/Mic-hackathon-2024-codes-publication/tree/1.0.0.1
Beyond Correlation: Towards Causal Large Language Model Agents in Biomedicine
May 22, 2025Abstract:Large Language Models (LLMs) show promise in biomedicine but lack true causal understanding, relying instead on correlations. This paper envisions causal LLM agents that integrate multimodal data (text, images, genomics, etc.) and perform intervention-based reasoning to infer cause-and-effect. Addressing this requires overcoming key challenges: designing safe, controllable agentic frameworks; developing rigorous benchmarks for causal evaluation; integrating heterogeneous data sources; and synergistically combining LLMs with structured knowledge (KGs) and formal causal inference tools. Such agents could unlock transformative opportunities, including accelerating drug discovery through automated hypothesis generation and simulation, enabling personalized medicine through patient-specific causal models. This research agenda aims to foster interdisciplinary efforts, bridging causal concepts and foundation models to develop reliable AI partners for biomedical progress.
34 Examples of LLM Applications in Materials Science and Chemistry: Towards Automation, Assistants, Agents, and Accelerated Scientific Discovery
May 05, 2025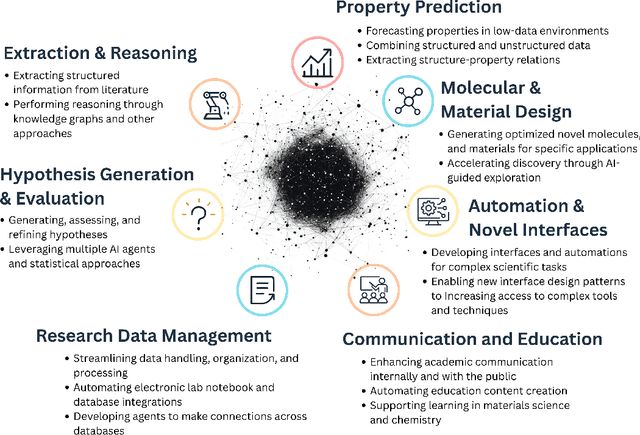
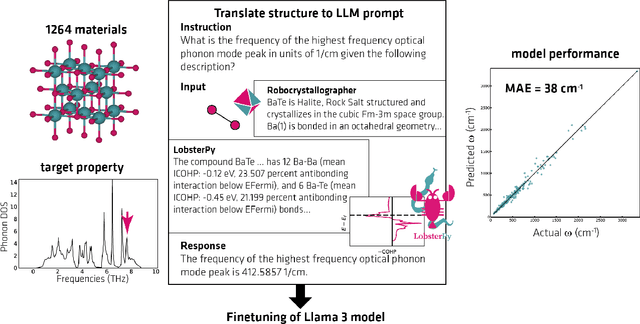
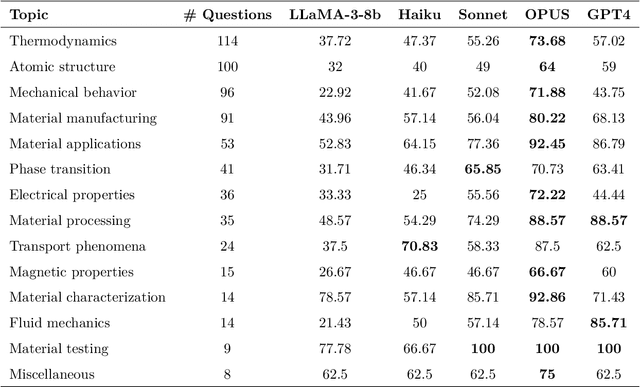
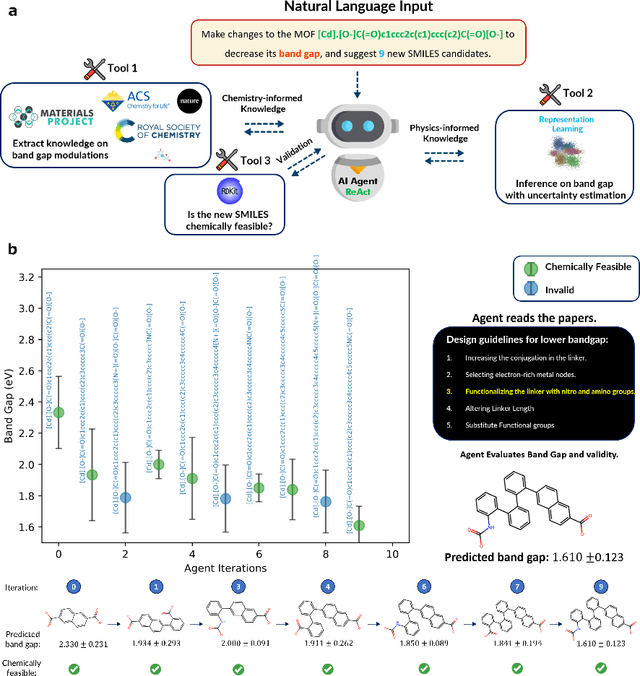
Abstract:Large Language Models (LLMs) are reshaping many aspects of materials science and chemistry research, enabling advances in molecular property prediction, materials design, scientific automation, knowledge extraction, and more. Recent developments demonstrate that the latest class of models are able to integrate structured and unstructured data, assist in hypothesis generation, and streamline research workflows. To explore the frontier of LLM capabilities across the research lifecycle, we review applications of LLMs through 34 total projects developed during the second annual Large Language Model Hackathon for Applications in Materials Science and Chemistry, a global hybrid event. These projects spanned seven key research areas: (1) molecular and material property prediction, (2) molecular and material design, (3) automation and novel interfaces, (4) scientific communication and education, (5) research data management and automation, (6) hypothesis generation and evaluation, and (7) knowledge extraction and reasoning from the scientific literature. Collectively, these applications illustrate how LLMs serve as versatile predictive models, platforms for rapid prototyping of domain-specific tools, and much more. In particular, improvements in both open source and proprietary LLM performance through the addition of reasoning, additional training data, and new techniques have expanded effectiveness, particularly in low-data environments and interdisciplinary research. As LLMs continue to improve, their integration into scientific workflows presents both new opportunities and new challenges, requiring ongoing exploration, continued refinement, and further research to address reliability, interpretability, and reproducibility.
Reflections from the 2024 Large Language Model (LLM) Hackathon for Applications in Materials Science and Chemistry
Nov 20, 2024
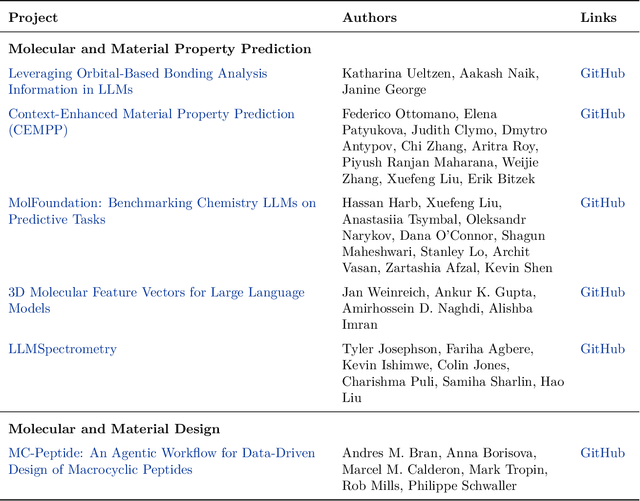
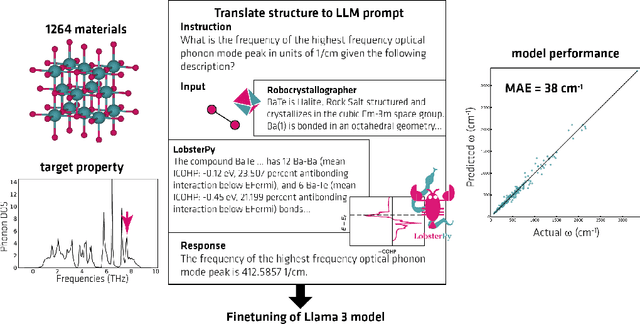
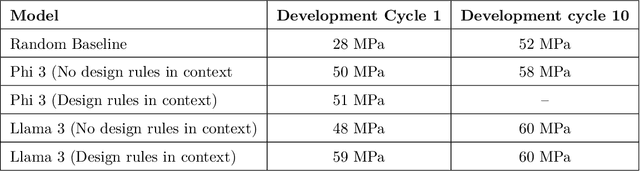
Abstract:Here, we present the outcomes from the second Large Language Model (LLM) Hackathon for Applications in Materials Science and Chemistry, which engaged participants across global hybrid locations, resulting in 34 team submissions. The submissions spanned seven key application areas and demonstrated the diverse utility of LLMs for applications in (1) molecular and material property prediction; (2) molecular and material design; (3) automation and novel interfaces; (4) scientific communication and education; (5) research data management and automation; (6) hypothesis generation and evaluation; and (7) knowledge extraction and reasoning from scientific literature. Each team submission is presented in a summary table with links to the code and as brief papers in the appendix. Beyond team results, we discuss the hackathon event and its hybrid format, which included physical hubs in Toronto, Montreal, San Francisco, Berlin, Lausanne, and Tokyo, alongside a global online hub to enable local and virtual collaboration. Overall, the event highlighted significant improvements in LLM capabilities since the previous year's hackathon, suggesting continued expansion of LLMs for applications in materials science and chemistry research. These outcomes demonstrate the dual utility of LLMs as both multipurpose models for diverse machine learning tasks and platforms for rapid prototyping custom applications in scientific research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge