AI- and HPC-enabled Lead Generation for SARS-CoV-2: Models and Processes to Extract Druglike Molecules Contained in Natural Language Text
Paper and Code
Jan 12, 2021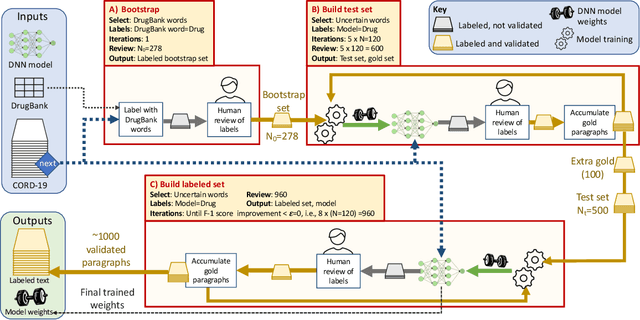
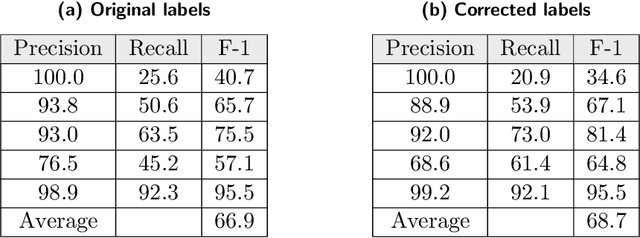
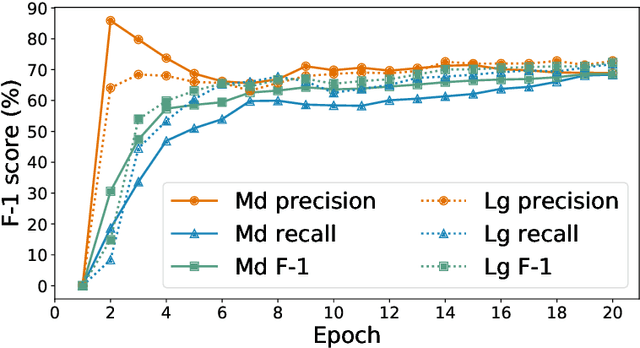
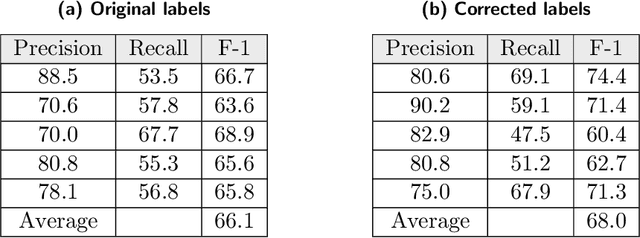
Researchers worldwide are seeking to repurpose existing drugs or discover new drugs to counter the disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A promising source of candidates for such studies is molecules that have been reported in the scientific literature to be drug-like in the context of coronavirus research. We report here on a project that leverages both human and artificial intelligence to detect references to drug-like molecules in free text. We engage non-expert humans to create a corpus of labeled text, use this labeled corpus to train a named entity recognition model, and employ the trained model to extract 10912 drug-like molecules from the COVID-19 Open Research Dataset Challenge (CORD-19) corpus of 198875 papers. Performance analyses show that our automated extraction model can achieve performance on par with that of non-expert humans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge