Neal Yuan
EchoPrime: A Multi-Video View-Informed Vision-Language Model for Comprehensive Echocardiography Interpretation
Oct 13, 2024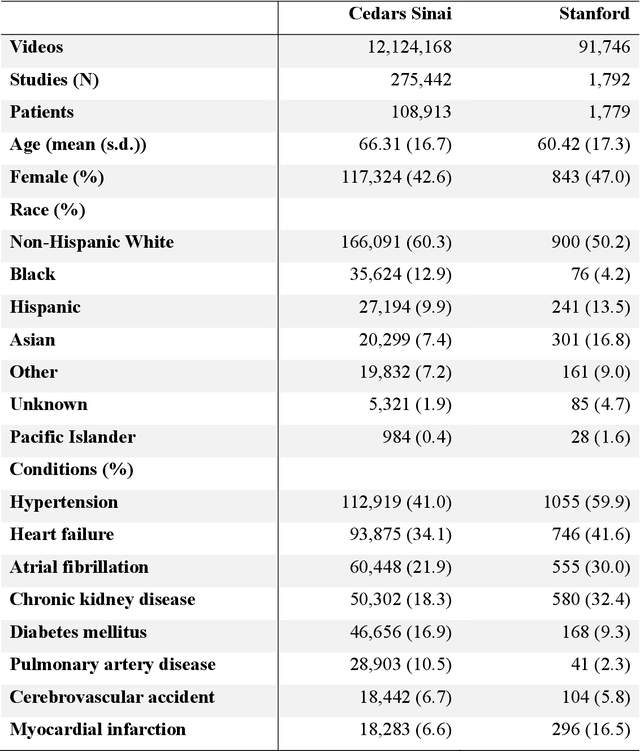
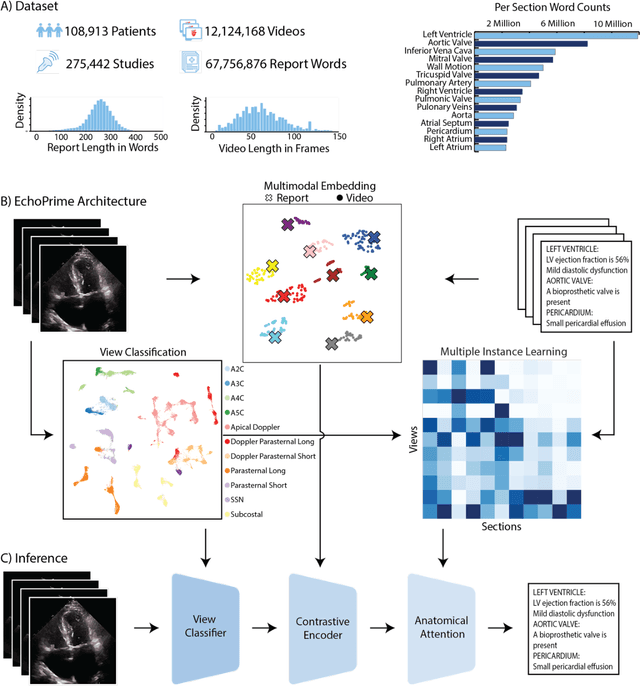
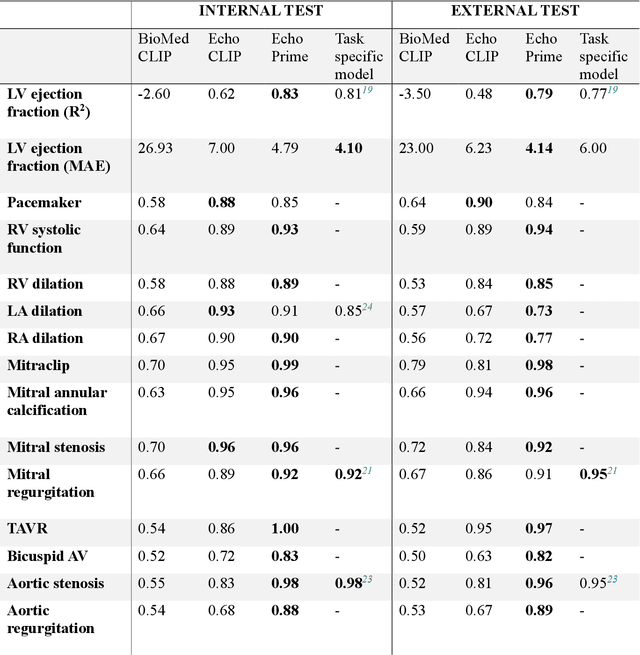
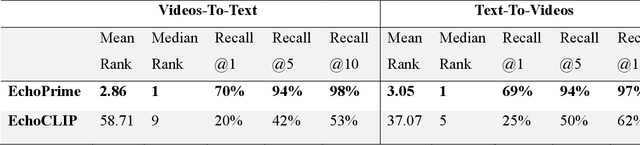
Abstract:Echocardiography is the most widely used cardiac imaging modality, capturing ultrasound video data to assess cardiac structure and function. Artificial intelligence (AI) in echocardiography has the potential to streamline manual tasks and improve reproducibility and precision. However, most echocardiography AI models are single-view, single-task systems that do not synthesize complementary information from multiple views captured during a full exam, and thus lead to limited performance and scope of applications. To address this problem, we introduce EchoPrime, a multi-view, view-informed, video-based vision-language foundation model trained on over 12 million video-report pairs. EchoPrime uses contrastive learning to train a unified embedding model for all standard views in a comprehensive echocardiogram study with representation of both rare and common diseases and diagnoses. EchoPrime then utilizes view-classification and a view-informed anatomic attention model to weight video-specific interpretations that accurately maps the relationship between echocardiographic views and anatomical structures. With retrieval-augmented interpretation, EchoPrime integrates information from all echocardiogram videos in a comprehensive study and performs holistic comprehensive clinical echocardiography interpretation. In datasets from two independent healthcare systems, EchoPrime achieves state-of-the art performance on 23 diverse benchmarks of cardiac form and function, surpassing the performance of both task-specific approaches and prior foundation models. Following rigorous clinical evaluation, EchoPrime can assist physicians in the automated preliminary assessment of comprehensive echocardiography.
Multimodal Foundation Models For Echocardiogram Interpretation
Sep 02, 2023Abstract:Multimodal deep learning foundation models can learn the relationship between images and text. In the context of medical imaging, mapping images to language concepts reflects the clinical task of diagnostic image interpretation, however current general-purpose foundation models do not perform well in this context because their training corpus have limited medical text and images. To address this challenge and account for the range of cardiac physiology, we leverage 1,032,975 cardiac ultrasound videos and corresponding expert interpretations to develop EchoCLIP, a multimodal foundation model for echocardiography. EchoCLIP displays strong zero-shot (not explicitly trained) performance in cardiac function assessment (external validation left ventricular ejection fraction mean absolute error (MAE) of 7.1%) and identification of implanted intracardiac devices (areas under the curve (AUC) between 0.84 and 0.98 for pacemakers and artificial heart valves). We also developed a long-context variant (EchoCLIP-R) with a custom echocardiography report text tokenizer which can accurately identify unique patients across multiple videos (AUC of 0.86), identify clinical changes such as orthotopic heart transplants (AUC of 0.79) or cardiac surgery (AUC 0.77), and enable robust image-to-text search (mean cross-modal retrieval rank in the top 1% of candidate text reports). These emergent capabilities can be used for preliminary assessment and summarization of echocardiographic findings.
Deep Learning Discovery of Demographic Biomarkers in Echocardiography
Jul 13, 2022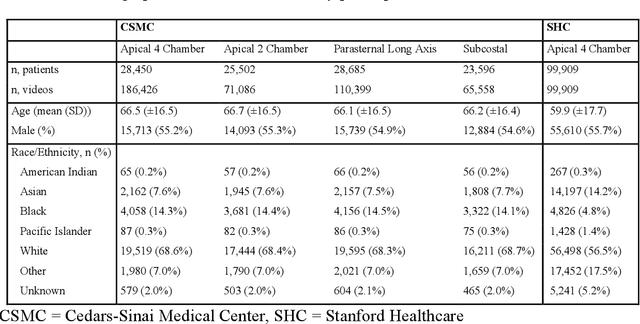
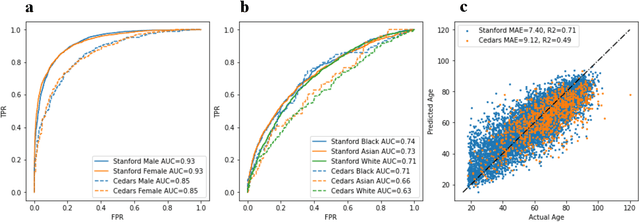
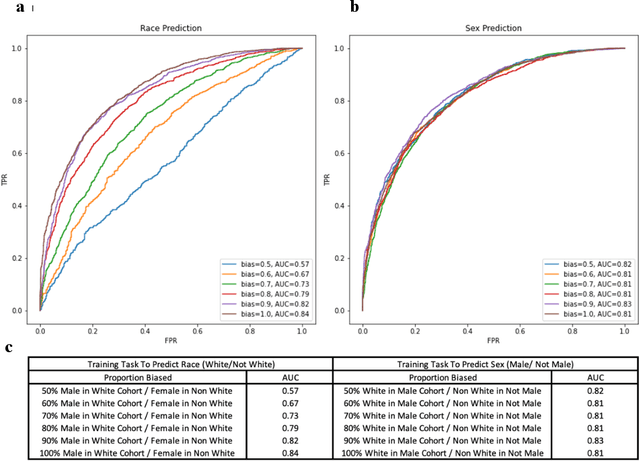
Abstract:Deep learning has been shown to accurately assess 'hidden' phenotypes and predict biomarkers from medical imaging beyond traditional clinician interpretation of medical imaging. Given the black box nature of artificial intelligence (AI) models, caution should be exercised in applying models to healthcare as prediction tasks might be short-cut by differences in demographics across disease and patient populations. Using large echocardiography datasets from two healthcare systems, we test whether it is possible to predict age, race, and sex from cardiac ultrasound images using deep learning algorithms and assess the impact of varying confounding variables. We trained video-based convolutional neural networks to predict age, sex, and race. We found that deep learning models were able to identify age and sex, while unable to reliably predict race. Without considering confounding differences between categories, the AI model predicted sex with an AUC of 0.85 (95% CI 0.84 - 0.86), age with a mean absolute error of 9.12 years (95% CI 9.00 - 9.25), and race with AUCs ranging from 0.63 - 0.71. When predicting race, we show that tuning the proportion of a confounding variable (sex) in the training data significantly impacts model AUC (ranging from 0.57 to 0.84), while in training a sex prediction model, tuning a confounder (race) did not substantially change AUC (0.81 - 0.83). This suggests a significant proportion of the model's performance on predicting race could come from confounding features being detected by AI. Further work remains to identify the particular imaging features that associate with demographic information and to better understand the risks of demographic identification in medical AI as it pertains to potentially perpetuating bias and disparities.
High-Throughput Precision Phenotyping of Left Ventricular Hypertrophy with Cardiovascular Deep Learning
Jun 23, 2021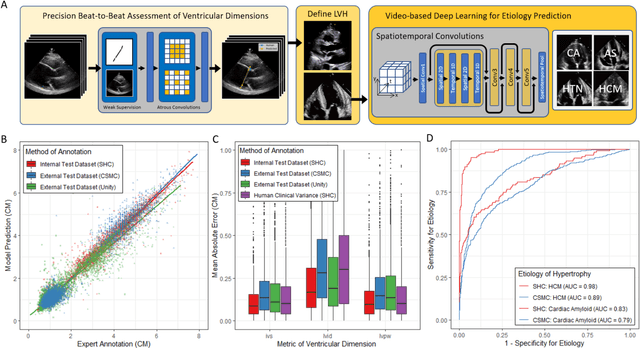
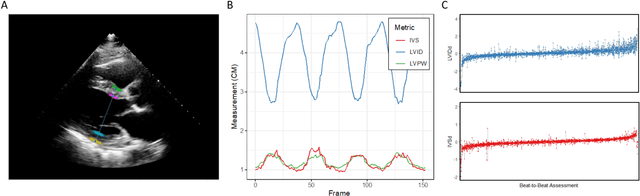
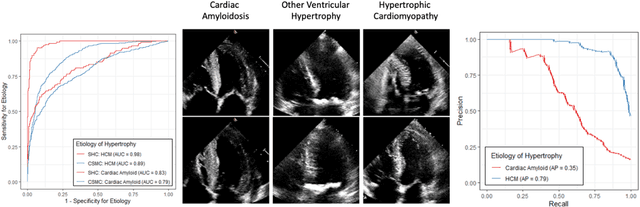
Abstract:Left ventricular hypertrophy (LVH) results from chronic remodeling caused by a broad range of systemic and cardiovascular disease including hypertension, aortic stenosis, hypertrophic cardiomyopathy, and cardiac amyloidosis. Early detection and characterization of LVH can significantly impact patient care but is limited by under-recognition of hypertrophy, measurement error and variability, and difficulty differentiating etiologies of LVH. To overcome this challenge, we present EchoNet-LVH - a deep learning workflow that automatically quantifies ventricular hypertrophy with precision equal to human experts and predicts etiology of LVH. Trained on 28,201 echocardiogram videos, our model accurately measures intraventricular wall thickness (mean absolute error [MAE] 1.4mm, 95% CI 1.2-1.5mm), left ventricular diameter (MAE 2.4mm, 95% CI 2.2-2.6mm), and posterior wall thickness (MAE 1.2mm, 95% CI 1.1-1.3mm) and classifies cardiac amyloidosis (area under the curve of 0.83) and hypertrophic cardiomyopathy (AUC 0.98) from other etiologies of LVH. In external datasets from independent domestic and international healthcare systems, EchoNet-LVH accurately quantified ventricular parameters (R2 of 0.96 and 0.90 respectively) and detected cardiac amyloidosis (AUC 0.79) and hypertrophic cardiomyopathy (AUC 0.89) on the domestic external validation site. Leveraging measurements across multiple heart beats, our model can more accurately identify subtle changes in LV geometry and its causal etiologies. Compared to human experts, EchoNet-LVH is fully automated, allowing for reproducible, precise measurements, and lays the foundation for precision diagnosis of cardiac hypertrophy. As a resource to promote further innovation, we also make publicly available a large dataset of 23,212 annotated echocardiogram videos.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge