Adolf Pfefferbaum
Self-Supervised Longitudinal Neighbourhood Embedding
Mar 09, 2021



Abstract:Longitudinal MRIs are often used to capture the gradual deterioration of brain structure and function caused by aging or neurological diseases. Analyzing this data via machine learning generally requires a large number of ground-truth labels, which are often missing or expensive to obtain. Reducing the need for labels, we propose a self-supervised strategy for representation learning named Longitudinal Neighborhood Embedding (LNE). Motivated by concepts in contrastive learning, LNE explicitly models the similarity between trajectory vectors across different subjects. We do so by building a graph in each training iteration defining neighborhoods in the latent space so that the progression direction of a subject follows the direction of its neighbors. This results in a smooth trajectory field that captures the global morphological change of the brain while maintaining the local continuity. We apply LNE to longitudinal T1w MRIs of two neuroimaging studies: a dataset composed of 274 healthy subjects, and Alzheimer's Disease Neuroimaging Initiative (ADNI, N=632). The visualization of the smooth trajectory vector field and superior performance on downstream tasks demonstrate the strength of the proposed method over existing self-supervised methods in extracting information associated with normal aging and in revealing the impact of neurodegenerative disorders. The code is available at \url{https://github.com/ouyangjiahong/longitudinal-neighbourhood-embedding.git}.
Vision-based Estimation of MDS-UPDRS Gait Scores for Assessing Parkinson's Disease Motor Severity
Jul 17, 2020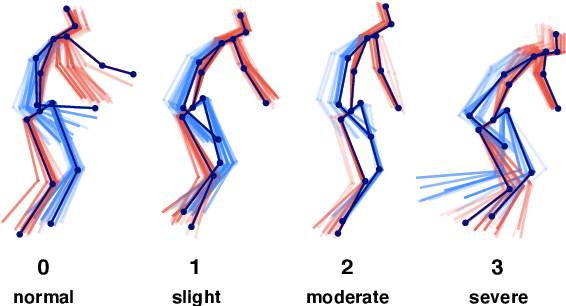
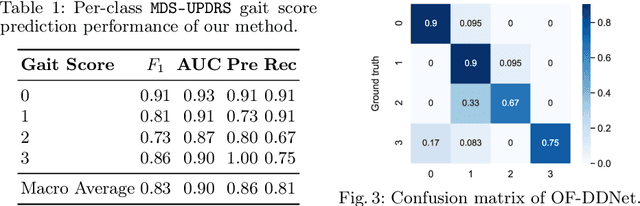
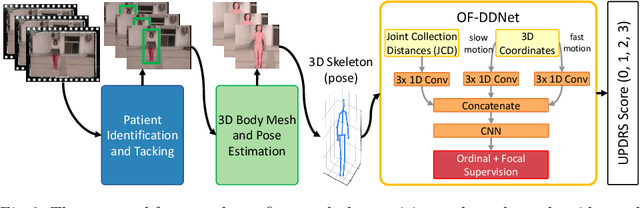
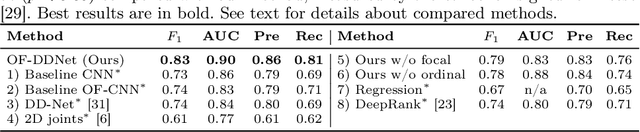
Abstract:Parkinson's disease (PD) is a progressive neurological disorder primarily affecting motor function resulting in tremor at rest, rigidity, bradykinesia, and postural instability. The physical severity of PD impairments can be quantified through the Movement Disorder Society Unified Parkinson's Disease Rating Scale (MDS-UPDRS), a widely used clinical rating scale. Accurate and quantitative assessment of disease progression is critical to developing a treatment that slows or stops further advancement of the disease. Prior work has mainly focused on dopamine transport neuroimaging for diagnosis or costly and intrusive wearables evaluating motor impairments. For the first time, we propose a computer vision-based model that observes non-intrusive video recordings of individuals, extracts their 3D body skeletons, tracks them through time, and classifies the movements according to the MDS-UPDRS gait scores. Experimental results show that our proposed method performs significantly better than chance and competing methods with an F1-score of 0.83 and a balanced accuracy of 81%. This is the first benchmark for classifying PD patients based on MDS-UPDRS gait severity and could be an objective biomarker for disease severity. Our work demonstrates how computer-assisted technologies can be used to non-intrusively monitor patients and their motor impairments. The code is available at https://github.com/mlu355/PD-Motor-Severity-Estimation.
Recurrent Neural Networks with Longitudinal Pooling and Consistency Regularization
Mar 31, 2020



Abstract:Most neurological diseases are characterized by gradual deterioration of brain structure and function. To identify the impact of such diseases, studies have been acquiring large longitudinal MRI datasets and applied deep-learning to predict diagnosis label(s). These learning models apply Convolutional Neural Networks (CNN) to extract informative features from each time point of the longitudinal MRI and Recurrent Neural Networks (RNN) to classify each time point based on those features. However, they neglect the progressive nature of the disease, which may result in clinically implausible predictions across visits. In this paper, we propose a framework that injects the extracted features from CNNs at each time point to the RNN cells considering the dependencies across different time points in the longitudinal data. On the feature level, we propose a novel longitudinal pooling layer to couple features of a visit with those of proceeding ones. On the prediction level, we add a consistency regularization to the classification objective in line with the nature of the disease progression across visits. We evaluate the proposed method on the longitudinal structural MRIs from three neuroimaging datasets: Alzheimer's Disease Neuroimaging Initiative (ADNI, N=404), a dataset composed of 274 healthy controls and 329 patients with Alcohol Use Disorder (AUD), and 255 youths from the National Consortium on Alcohol and NeuroDevelopment in Adolescence (NCANDA). All three experiments show that our method is superior to the widely used methods. The code is available at https://github.com/ouyangjiahong/longitudinal-pooling.
Spatio-Temporal Graph Convolution for Functional MRI Analysis
Mar 24, 2020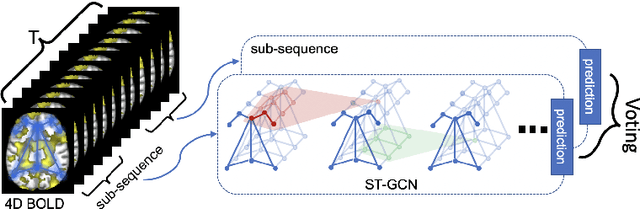
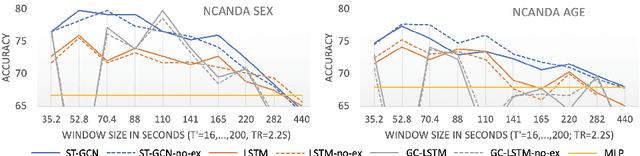
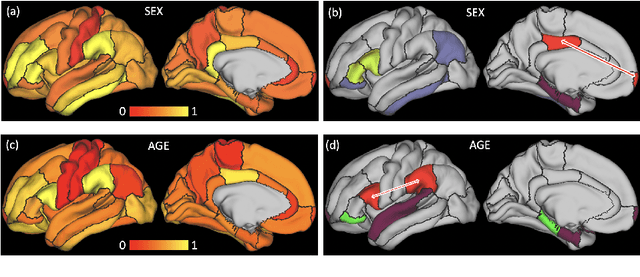
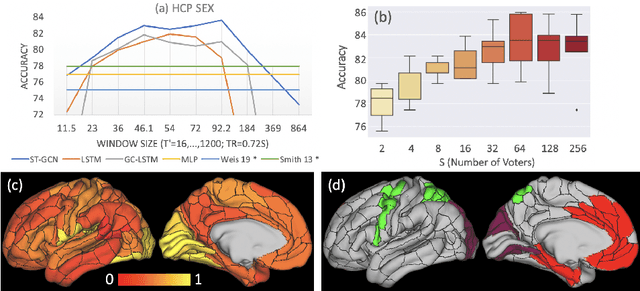
Abstract:The BOLD signal of resting-state fMRI (rs-fMRI) records the functional brain connectivity in a rich dynamic spatio-temporal setting. However, existing methods applied to rs-fMRI often fail to consider both spatial and temporal characteristics of the data. They either neglect the functional dependency between different brain regions in a network or discard the information in the temporal dynamics of brain activity. To overcome those shortcomings, we propose to formulate functional connectivity networks within the context of spatio-temporal graphs. We then train a spatio-temporal graph convolutional network (ST-GCN) on short sub-sequences of the BOLD time series to model the non-stationary nature of functional connectivity. We simultaneously learn the graph edge importance within ST-GCN to enable interpretation of functional connectivities contributing to the prediction model. In analyzing the rs-fMRI of the Human Connectome Project (HCP, N=1,091) and the National Consortium on Alcohol and Neurodevelopment in Adolescence (NCANDA, N=773), ST-GCN is significantly more accurate than common approaches in predicting gender and age based on BOLD signals. The matrix recording edge importance localizes brain regions and functional connections with significant aging and sex effects, which are verified by the neuroscience literature.
Bias-Resilient Neural Network
Oct 08, 2019
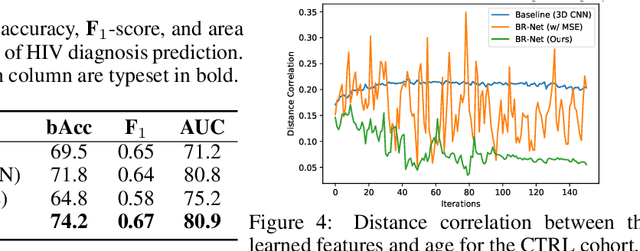
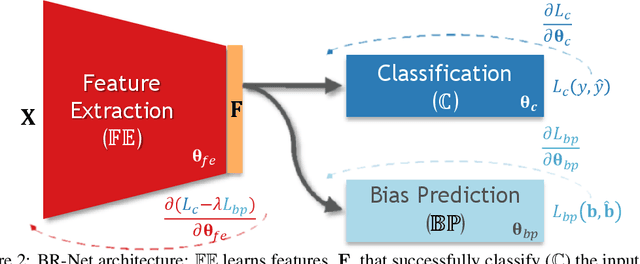
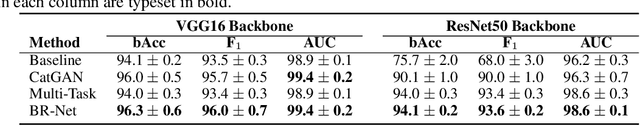
Abstract:Presence of bias and confounding effects is inarguably one of the most critical challenges in machine learning applications that has alluded to pivotal debates in the recent years. Such challenges range from spurious associations of confounding variables in medical studies to the bias of race in gender or face recognition systems. One solution is to enhance datasets and organize them such that they do not reflect biases, which is a cumbersome and intensive task. The alternative is to make use of available data and build models considering these biases. Traditional statistical methods apply straightforward techniques such as residualization or stratification to precomputed features to account for confounding variables. However, these techniques are generally not suitable for end-to-end deep learning methods. In this paper, we propose a method based on the adversarial training strategy to learn discriminative features unbiased and invariant to the confounder(s). This is enabled by incorporating a new adversarial loss function that encourages a vanished correlation between the bias and learned features. We apply our method to synthetic data, medical images, and a gender classification (Gender Shades Pilot Parliaments Benchmark) dataset. Our results show that the learned features by our method not only result in superior prediction performance but also are uncorrelated with the bias or confounder variables. The code is available at http://github.com/QingyuZhao/BR-Net/.
Confounder-Aware Visualization of ConvNets
Jul 30, 2019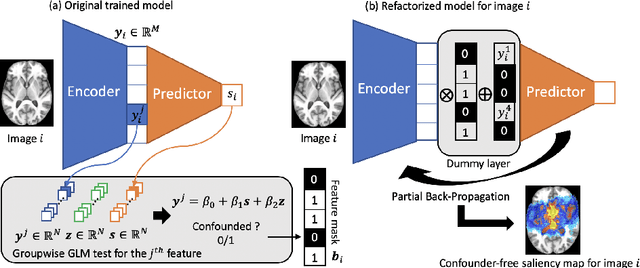
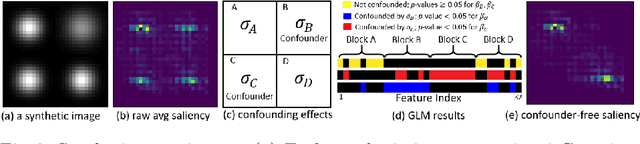
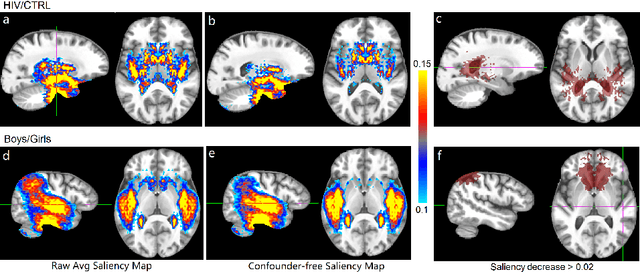
Abstract:With recent advances in deep learning, neuroimaging studies increasingly rely on convolutional networks (ConvNets) to predict diagnosis based on MR images. To gain a better understanding of how a disease impacts the brain, the studies visualize the salience maps of the ConvNet highlighting voxels within the brain majorly contributing to the prediction. However, these salience maps are generally confounded, i.e., some salient regions are more predictive of confounding variables (such as age) than the diagnosis. To avoid such misinterpretation, we propose in this paper an approach that aims to visualize confounder-free saliency maps that only highlight voxels predictive of the diagnosis. The approach incorporates univariate statistical tests to identify confounding effects within the intermediate features learned by ConvNet. The influence from the subset of confounded features is then removed by a novel partial back-propagation procedure. We use this two-step approach to visualize confounder-free saliency maps extracted from synthetic and two real datasets. These experiments reveal the potential of our visualization in producing unbiased model-interpretation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge