Akshay S Chaudhari
Preference Fine-Tuning for Factuality in Chest X-Ray Interpretation Models Without Human Feedback
Oct 09, 2024



Abstract:Radiologists play a crucial role by translating medical images into medical reports. However, the field faces staffing shortages and increasing workloads. While automated approaches using vision-language models (VLMs) show promise as assistants, they require exceptionally high accuracy. Most current VLMs in radiology rely solely on supervised fine-tuning (SFT). Meanwhile, in the general domain, additional preference fine-tuning has become standard practice. The challenge in radiology lies in the prohibitive cost of obtaining radiologist feedback. We propose a scalable automated preference alignment technique for VLMs in radiology, focusing on chest X-ray (CXR) report generation. Our method leverages publicly available datasets with an LLM-as-a-Judge mechanism, eliminating the need for additional expert radiologist feedback. We evaluate and benchmark five direct alignment algorithms (DAAs). Our results show up to a 57.4% improvement in average GREEN scores, a LLM-based metric for evaluating CXR reports, and a 9.2% increase in an average across six metrics (domain specific and general), compared to the SFT baseline. We study reward overoptimization via length exploitation, with reports lengthening by up to 3.2x. To assess a potential alignment tax, we benchmark on six additional diverse tasks, finding no significant degradations. A reader study involving four board-certified radiologists indicates win rates of up to 0.62 over the SFT baseline, while significantly penalizing verbosity. Our analysis provides actionable insights for the development of VLMs in high-stakes fields like radiology.
GREEN: Generative Radiology Report Evaluation and Error Notation
May 06, 2024


Abstract:Evaluating radiology reports is a challenging problem as factual correctness is extremely important due to the need for accurate medical communication about medical images. Existing automatic evaluation metrics either suffer from failing to consider factual correctness (e.g., BLEU and ROUGE) or are limited in their interpretability (e.g., F1CheXpert and F1RadGraph). In this paper, we introduce GREEN (Generative Radiology Report Evaluation and Error Notation), a radiology report generation metric that leverages the natural language understanding of language models to identify and explain clinically significant errors in candidate reports, both quantitatively and qualitatively. Compared to current metrics, GREEN offers: 1) a score aligned with expert preferences, 2) human interpretable explanations of clinically significant errors, enabling feedback loops with end-users, and 3) a lightweight open-source method that reaches the performance of commercial counterparts. We validate our GREEN metric by comparing it to GPT-4, as well as to error counts of 6 experts and preferences of 2 experts. Our method demonstrates not only higher correlation with expert error counts, but simultaneously higher alignment with expert preferences when compared to previous approaches."
SKM-TEA: A Dataset for Accelerated MRI Reconstruction with Dense Image Labels for Quantitative Clinical Evaluation
Mar 14, 2022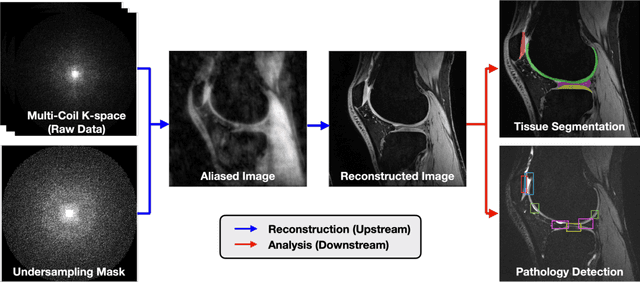
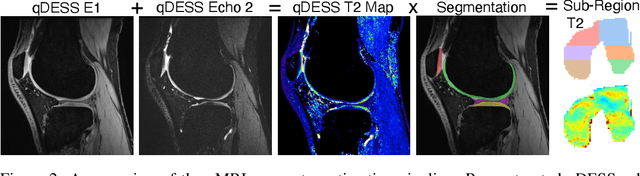
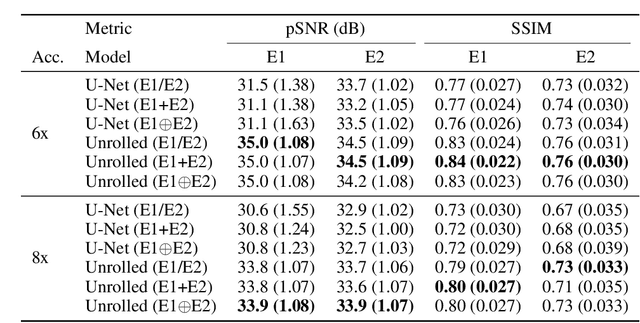
Abstract:Magnetic resonance imaging (MRI) is a cornerstone of modern medical imaging. However, long image acquisition times, the need for qualitative expert analysis, and the lack of (and difficulty extracting) quantitative indicators that are sensitive to tissue health have curtailed widespread clinical and research studies. While recent machine learning methods for MRI reconstruction and analysis have shown promise for reducing this burden, these techniques are primarily validated with imperfect image quality metrics, which are discordant with clinically-relevant measures that ultimately hamper clinical deployment and clinician trust. To mitigate this challenge, we present the Stanford Knee MRI with Multi-Task Evaluation (SKM-TEA) dataset, a collection of quantitative knee MRI (qMRI) scans that enables end-to-end, clinically-relevant evaluation of MRI reconstruction and analysis tools. This 1.6TB dataset consists of raw-data measurements of ~25,000 slices (155 patients) of anonymized patient MRI scans, the corresponding scanner-generated DICOM images, manual segmentations of four tissues, and bounding box annotations for sixteen clinically relevant pathologies. We provide a framework for using qMRI parameter maps, along with image reconstructions and dense image labels, for measuring the quality of qMRI biomarker estimates extracted from MRI reconstruction, segmentation, and detection techniques. Finally, we use this framework to benchmark state-of-the-art baselines on this dataset. We hope our SKM-TEA dataset and code can enable a broad spectrum of research for modular image reconstruction and image analysis in a clinically informed manner. Dataset access, code, and benchmarks are available at https://github.com/StanfordMIMI/skm-tea.
VORTEX: Physics-Driven Data Augmentations for Consistency Training for Robust Accelerated MRI Reconstruction
Nov 03, 2021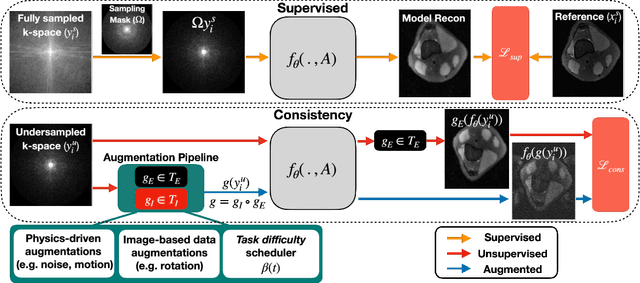
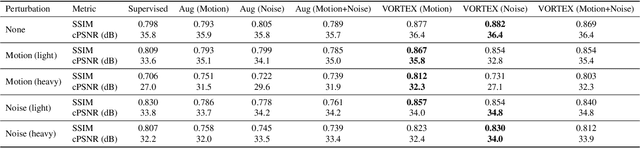
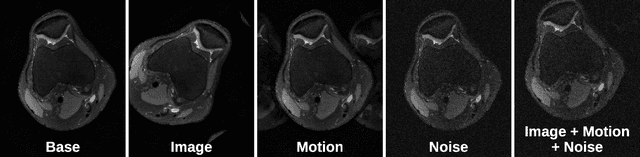
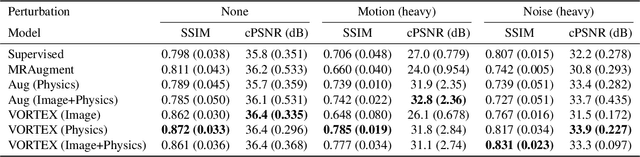
Abstract:Deep neural networks have enabled improved image quality and fast inference times for various inverse problems, including accelerated magnetic resonance imaging (MRI) reconstruction. However, such models require large amounts of fully-sampled ground truth data, which are difficult to curate and are sensitive to distribution drifts. In this work, we propose applying physics-driven data augmentations for consistency training that leverage our domain knowledge of the forward MRI data acquisition process and MRI physics for improved data efficiency and robustness to clinically-relevant distribution drifts. Our approach, termed VORTEX (1) demonstrates strong improvements over supervised baselines with and without augmentation in robustness to signal-to-noise ratio change and motion corruption in data-limited regimes; (2) considerably outperforms state-of-the-art data augmentation techniques that are purely image-based on both in-distribution and out-of-distribution data; and (3) enables composing heterogeneous image-based and physics-driven augmentations.
Noise2Recon: A Semi-Supervised Framework for Joint MRI Reconstruction and Denoising
Sep 30, 2021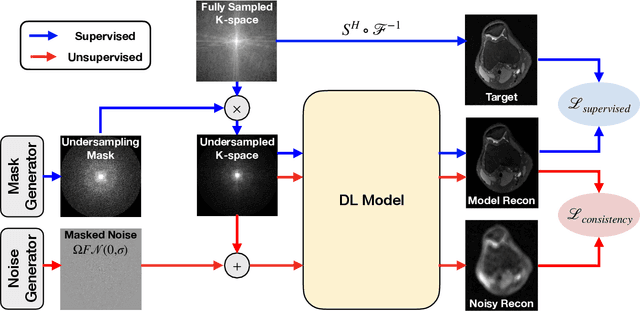
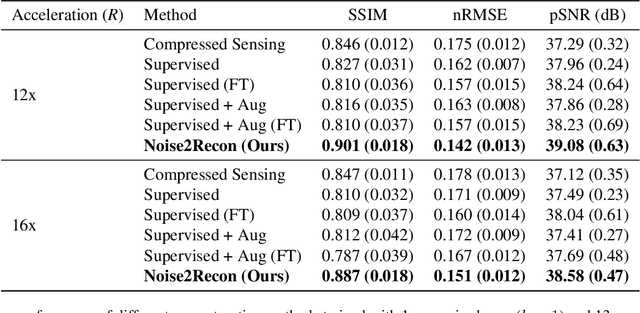
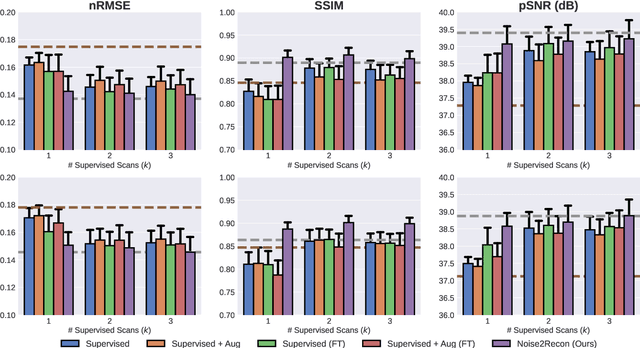
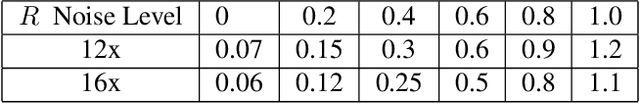
Abstract:Deep learning (DL) has shown promise for faster, high quality accelerated MRI reconstruction. However, standard supervised DL methods depend on extensive amounts of fully-sampled ground-truth data and are sensitive to out-of-distribution (OOD) shifts, in particular for low signal-to-noise ratio (SNR) acquisitions. To alleviate this challenge, we propose a semi-supervised, consistency-based framework (termed Noise2Recon) for joint MR reconstruction and denoising. Our method enables the usage of a limited number of fully-sampled and a large number of undersampled-only scans. We compare our method to augmentation-based supervised techniques and fine-tuned denoisers. Results demonstrate that even with minimal ground-truth data, Noise2Recon (1) achieves high performance on in-distribution (low-noise) scans and (2) improves generalizability to OOD, noisy scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge