Valentina Mazzoli
Deep Anatomical Federated Network (Dafne): an open client/server framework for the continuous collaborative improvement of deep-learning-based medical image segmentation
Feb 14, 2023Abstract:Semantic segmentation is a crucial step to extract quantitative information from medical (and, specifically, radiological) images to aid the diagnostic process, clinical follow-up. and to generate biomarkers for clinical research. In recent years, machine learning algorithms have become the primary tool for this task. However, its real-world performance is heavily reliant on the comprehensiveness of training data. Dafne is the first decentralized, collaborative solution that implements continuously evolving deep learning models exploiting the collective knowledge of the users of the system. In the Dafne workflow, the result of each automated segmentation is refined by the user through an integrated interface, so that the new information is used to continuously expand the training pool via federated incremental learning. The models deployed through Dafne are able to improve their performance over time and to generalize to data types not seen in the training sets, thus becoming a viable and practical solution for real-life medical segmentation tasks.
SKM-TEA: A Dataset for Accelerated MRI Reconstruction with Dense Image Labels for Quantitative Clinical Evaluation
Mar 14, 2022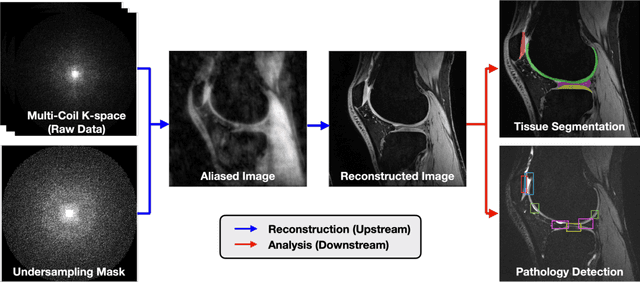
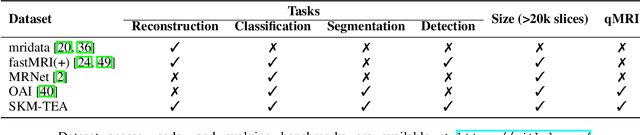
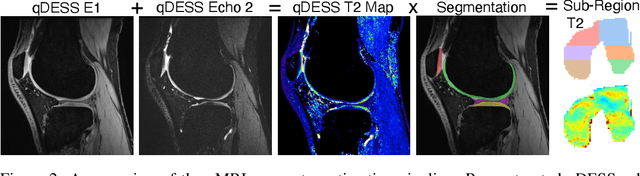
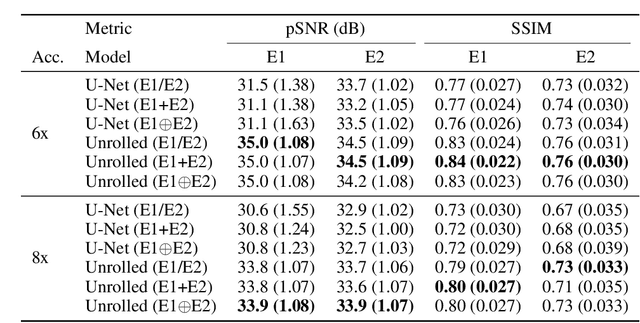
Abstract:Magnetic resonance imaging (MRI) is a cornerstone of modern medical imaging. However, long image acquisition times, the need for qualitative expert analysis, and the lack of (and difficulty extracting) quantitative indicators that are sensitive to tissue health have curtailed widespread clinical and research studies. While recent machine learning methods for MRI reconstruction and analysis have shown promise for reducing this burden, these techniques are primarily validated with imperfect image quality metrics, which are discordant with clinically-relevant measures that ultimately hamper clinical deployment and clinician trust. To mitigate this challenge, we present the Stanford Knee MRI with Multi-Task Evaluation (SKM-TEA) dataset, a collection of quantitative knee MRI (qMRI) scans that enables end-to-end, clinically-relevant evaluation of MRI reconstruction and analysis tools. This 1.6TB dataset consists of raw-data measurements of ~25,000 slices (155 patients) of anonymized patient MRI scans, the corresponding scanner-generated DICOM images, manual segmentations of four tissues, and bounding box annotations for sixteen clinically relevant pathologies. We provide a framework for using qMRI parameter maps, along with image reconstructions and dense image labels, for measuring the quality of qMRI biomarker estimates extracted from MRI reconstruction, segmentation, and detection techniques. Finally, we use this framework to benchmark state-of-the-art baselines on this dataset. We hope our SKM-TEA dataset and code can enable a broad spectrum of research for modular image reconstruction and image analysis in a clinically informed manner. Dataset access, code, and benchmarks are available at https://github.com/StanfordMIMI/skm-tea.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge