M. Brandon Westover
Massachusetts General Hospital, Boston, MA
Artificial Intelligence without Restriction Surpassing Human Intelligence with Probability One: Theoretical Insight into Secrets of the Brain with AI Twins of the Brain
Dec 04, 2024



Abstract:Artificial Intelligence (AI) has apparently become one of the most important techniques discovered by humans in history while the human brain is widely recognized as one of the most complex systems in the universe. One fundamental critical question which would affect human sustainability remains open: Will artificial intelligence (AI) evolve to surpass human intelligence in the future? This paper shows that in theory new AI twins with fresh cellular level of AI techniques for neuroscience could approximate the brain and its functioning systems (e.g. perception and cognition functions) with any expected small error and AI without restrictions could surpass human intelligence with probability one in the end. This paper indirectly proves the validity of the conjecture made by Frank Rosenblatt 70 years ago about the potential capabilities of AI, especially in the realm of artificial neural networks. Intelligence is just one of fortuitous but sophisticated creations of the nature which has not been fully discovered. Like mathematics and physics, with no restrictions artificial intelligence would lead to a new subject with its self-contained systems and principles. We anticipate that this paper opens new doors for 1) AI twins and other AI techniques to be used in cellular level of efficient neuroscience dynamic analysis, functioning analysis of the brain and brain illness solutions; 2) new worldwide collaborative scheme for interdisciplinary teams concurrently working on and modelling different types of neurons and synapses and different level of functioning subsystems of the brain with AI techniques; 3) development of low energy of AI techniques with the aid of fundamental neuroscience properties; and 4) new controllable, explainable and safe AI techniques with reasoning capabilities of discovering principles in nature.
Annotation of Sleep Depth Index with Scalable Deep Learning Yields Novel Digital Biomarkers for Sleep Health
Jul 05, 2024

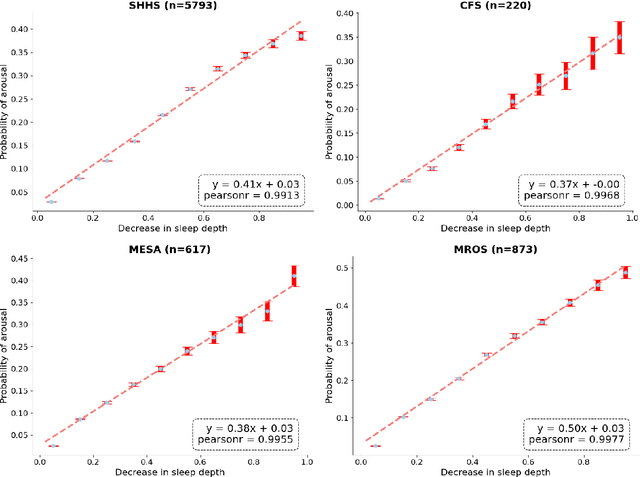
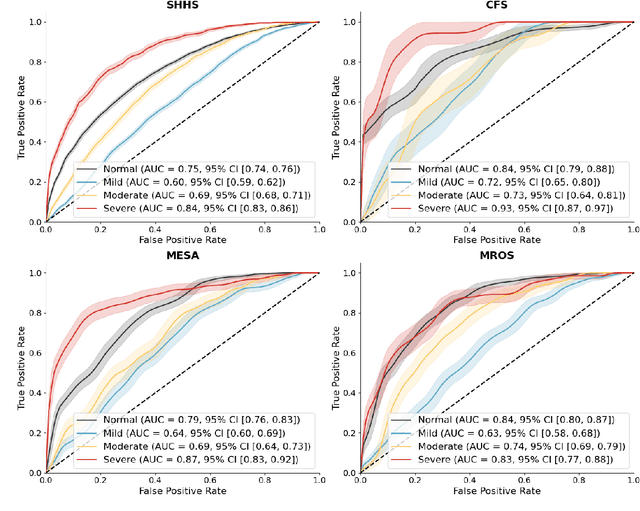
Abstract:Traditional sleep staging categorizes sleep and wakefulness into five coarse-grained classes, overlooking subtle variations within each stage. It provides limited information about the probability of arousal and may hinder the diagnosis of sleep disorders, such as insomnia. To address this issue, we propose a deep-learning method for automatic and scalable annotation of sleep depth index using existing sleep staging labels. Our approach is validated using polysomnography from over ten thousand recordings across four large-scale cohorts. The results show a strong correlation between the decrease in sleep depth index and the increase in arousal likelihood. Several case studies indicate that the sleep depth index captures more nuanced sleep structures than conventional sleep staging. Sleep biomarkers extracted from the whole-night sleep depth index exhibit statistically significant differences with medium-to-large effect sizes across groups of varied subjective sleep quality and insomnia symptoms. These sleep biomarkers also promise utility in predicting the severity of obstructive sleep apnea, particularly in severe cases. Our study underscores the utility of the proposed method for continuous sleep depth annotation, which could reveal more detailed structures and dynamics within whole-night sleep and yield novel digital biomarkers beneficial for sleep health.
Estimating Trustworthy and Safe Optimal Treatment Regimes
Oct 23, 2023



Abstract:Recent statistical and reinforcement learning methods have significantly advanced patient care strategies. However, these approaches face substantial challenges in high-stakes contexts, including missing data, inherent stochasticity, and the critical requirements for interpretability and patient safety. Our work operationalizes a safe and interpretable framework to identify optimal treatment regimes. This approach involves matching patients with similar medical and pharmacological characteristics, allowing us to construct an optimal policy via interpolation. We perform a comprehensive simulation study to demonstrate the framework's ability to identify optimal policies even in complex settings. Ultimately, we operationalize our approach to study regimes for treating seizures in critically ill patients. Our findings strongly support personalized treatment strategies based on a patient's medical history and pharmacological features. Notably, we identify that reducing medication doses for patients with mild and brief seizure episodes while adopting aggressive treatment for patients in intensive care unit experiencing intense seizures leads to more favorable outcomes.
BIOT: Cross-data Biosignal Learning in the Wild
May 10, 2023Abstract:Biological signals, such as electroencephalograms (EEG), play a crucial role in numerous clinical applications, exhibiting diverse data formats and quality profiles. Current deep learning models for biosignals are typically specialized for specific datasets and clinical settings, limiting their broader applicability. Motivated by the success of large language models in text processing, we explore the development of foundational models that are trained from multiple data sources and can be fine-tuned on different downstream biosignal tasks. To overcome the unique challenges associated with biosignals of various formats, such as mismatched channels, variable sample lengths, and prevalent missing values, we propose a Biosignal Transformer (\method). The proposed \method model can enable cross-data learning with mismatched channels, variable lengths, and missing values by tokenizing diverse biosignals into unified "biosignal sentences". Specifically, we tokenize each channel into fixed-length segments containing local signal features, flattening them to form consistent "sentences". Channel embeddings and {\em relative} position embeddings are added to preserve spatio-temporal features. The \method model is versatile and applicable to various biosignal learning settings across different datasets, including joint pre-training for larger models. Comprehensive evaluations on EEG, electrocardiogram (ECG), and human activity sensory signals demonstrate that \method outperforms robust baselines in common settings and facilitates learning across multiple datasets with different formats. Use CHB-MIT seizure detection task as an example, our vanilla \method model shows 3\% improvement over baselines in balanced accuracy, and the pre-trained \method models (optimized from other data sources) can further bring up to 4\% improvements.
Mapping the Ictal-Interictal-Injury Continuum Using Interpretable Machine Learning
Nov 14, 2022


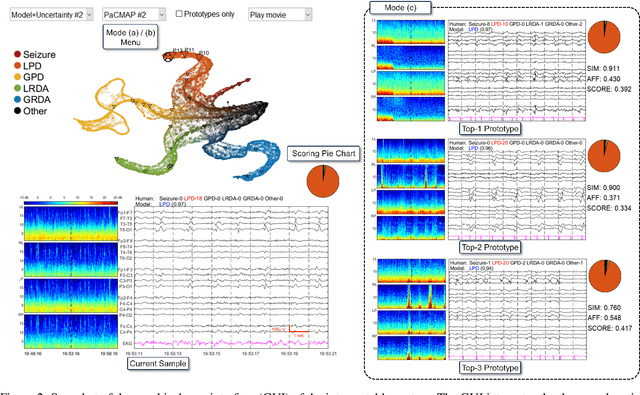
Abstract:IMPORTANCE: An interpretable machine learning model can provide faithful explanations of each prediction and yet maintain higher performance than its black box counterpart. OBJECTIVE: To design an interpretable machine learning model which accurately predicts EEG protopatterns while providing an explanation of its predictions with assistance of a specialized GUI. To map the cEEG latent features to a 2D space in order to visualize the ictal-interictal-injury continuum and gain insight into its high-dimensional structure. DESIGN, SETTING, AND PARTICIPANTS: 50,697 50-second cEEG samples from 2,711 ICU patients collected between July 2006 and March 2020 at Massachusetts General Hospital. Samples were labeled as one of 6 EEG activities by domain experts, with 124 different experts providing annotations. MAIN OUTCOMES AND MEASURES: Our neural network is interpretable because it uses case-based reasoning: it compares a new EEG reading to a set of learned prototypical EEG samples from the training dataset. Interpretability was measured with task-specific neighborhood agreement statistics. Discriminatory performance was evaluated with AUROC and AUPRC. RESULTS: The model achieves AUROCs of 0.87, 0.93, 0.96, 0.92, 0.93, 0.80 for classes Seizure, LPD, GPD, LRDA, GRDA, Other respectively. This performance is statistically significantly higher than that of the corresponding uninterpretable (black box) model with p<0.0001. Videos of the ictal-interictal-injury continuum are provided. CONCLUSION AND RELEVANCE: Our interpretable model and GUI can act as a reference for practitioners who work with cEEG patterns. We can now better understand the relationships between different types of cEEG patterns. In the future, this system may allow for targeted intervention and training in clinical settings. It could also be used for re-confirming or providing additional information for diagnostics.
Why Interpretable Causal Inference is Important for High-Stakes Decision Making for Critically Ill Patients and How To Do It
Mar 09, 2022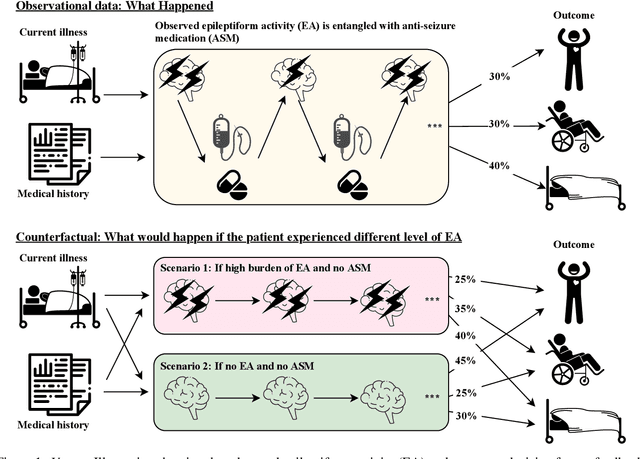
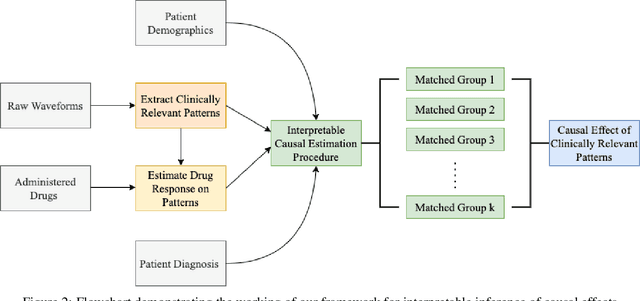
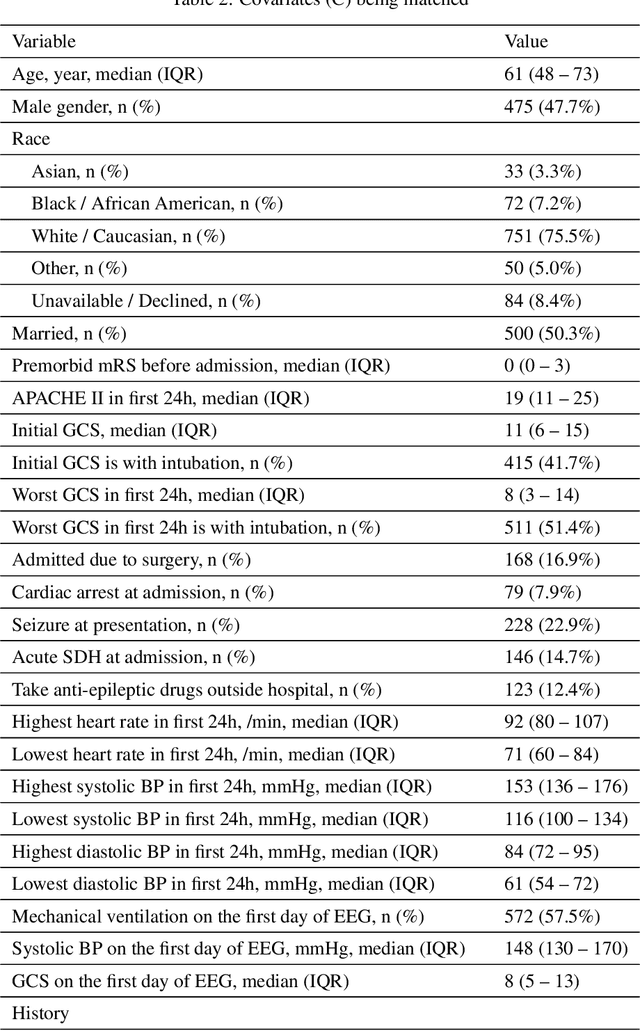
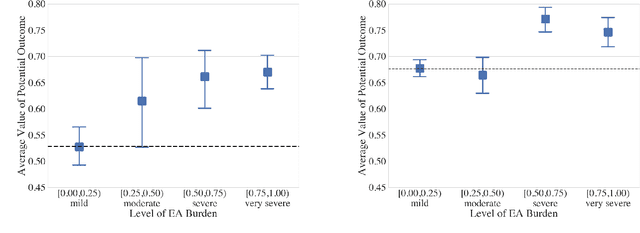
Abstract:Many fundamental problems affecting the care of critically ill patients lead to similar analytical challenges: physicians cannot easily estimate the effects of at-risk medical conditions or treatments because the causal effects of medical conditions and drugs are entangled. They also cannot easily perform studies: there are not enough high-quality data for high-dimensional observational causal inference, and RCTs often cannot ethically be conducted. However, mechanistic knowledge is available, including how drugs are absorbed into the body, and the combination of this knowledge with the limited data could potentially suffice -- if we knew how to combine them. In this work, we present a framework for interpretable estimation of causal effects for critically ill patients under exactly these complex conditions: interactions between drugs and observations over time, patient data sets that are not large, and mechanistic knowledge that can substitute for lack of data. We apply this framework to an extremely important problem affecting critically ill patients, namely the effect of seizures and other potentially harmful electrical events in the brain (called epileptiform activity -- EA) on outcomes. Given the high stakes involved and the high noise in the data, interpretability is critical for troubleshooting such complex problems. Interpretability of our matched groups allowed neurologists to perform chart reviews to verify the quality of our causal analysis. For instance, our work indicates that a patient who experiences a high level of seizure-like activity (75% high EA burden) and is untreated for a six-hour window, has, on average, a 16.7% increased chance of adverse outcomes such as severe brain damage, lifetime disability, or death. We find that patients with mild but long-lasting EA (average EA burden >= 50%) have their risk of an adverse outcome increased by 11.2%.
Using Deep Learning to Identify Patients with Cognitive Impairment in Electronic Health Records
Nov 13, 2021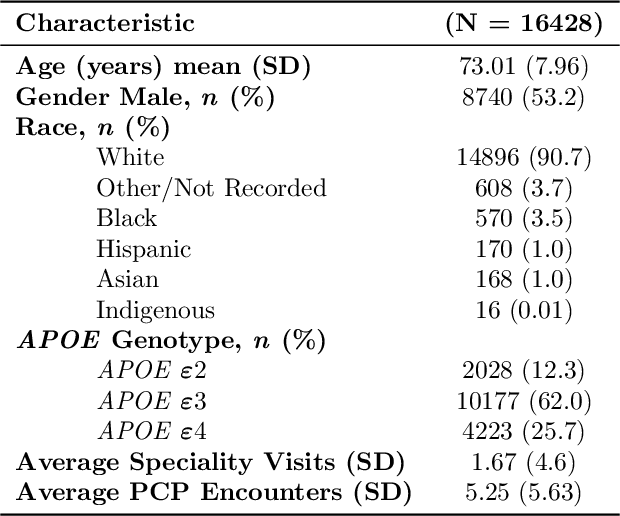


Abstract:Dementia is a neurodegenerative disorder that causes cognitive decline and affects more than 50 million people worldwide. Dementia is under-diagnosed by healthcare professionals - only one in four people who suffer from dementia are diagnosed. Even when a diagnosis is made, it may not be entered as a structured International Classification of Diseases (ICD) diagnosis code in a patient's charts. Information relevant to cognitive impairment (CI) is often found within electronic health records (EHR), but manual review of clinician notes by experts is both time consuming and often prone to errors. Automated mining of these notes presents an opportunity to label patients with cognitive impairment in EHR data. We developed natural language processing (NLP) tools to identify patients with cognitive impairment and demonstrate that linguistic context enhances performance for the cognitive impairment classification task. We fine-tuned our attention based deep learning model, which can learn from complex language structures, and substantially improved accuracy (0.93) relative to a baseline NLP model (0.84). Further, we show that deep learning NLP can successfully identify dementia patients without dementia-related ICD codes or medications.
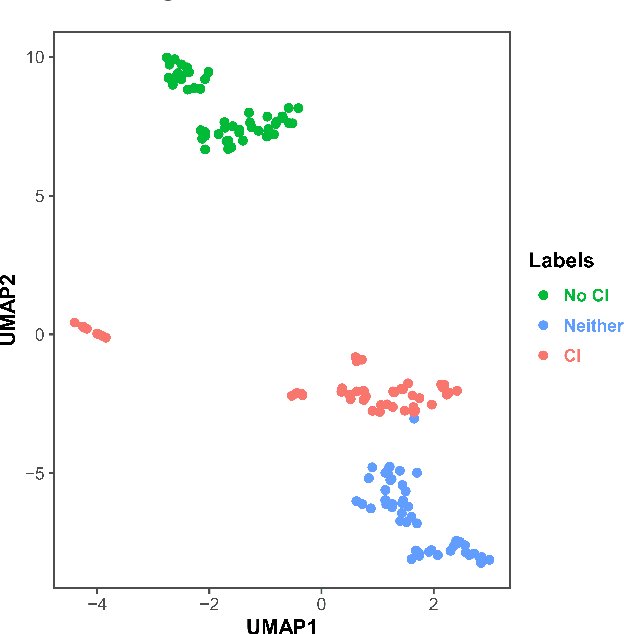
Self-supervised EEG Representation Learning for Automatic Sleep Staging
Oct 27, 2021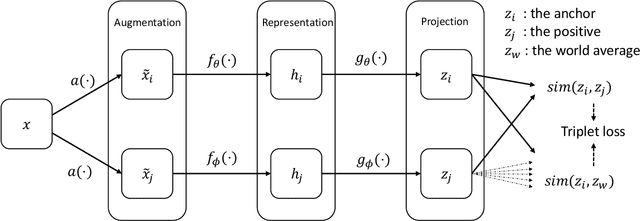
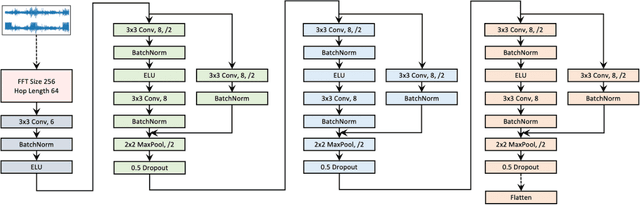
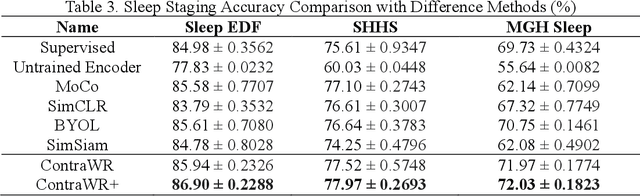
Abstract:Objective: In this paper, we aim to learn robust vector representations from massive unlabeled Electroencephalogram (EEG) signals, such that the learned representations (1) are expressive enough to replace the raw signals in the sleep staging task; and (2) provide better predictive performance than supervised models in scenarios of fewer labels and noisy samples. Materials and Methods: We propose a self-supervised model, named Contrast with the World Representation (ContraWR), for EEG signal representation learning, which uses global statistics from the dataset to distinguish signals associated with different sleep stages. The ContraWR model is evaluated on three real-world EEG datasets that include both at-home and in-lab recording settings. Results: ContraWR outperforms recent self-supervised learning methods, MoCo, SimCLR, BYOL, SimSiam on the sleep staging task across three datasets. ContraWR also beats supervised learning when fewer training labels are available (e.g., 4% accuracy improvement when less than 2% data is labeled). Moreover, the model provides informative representations in 2D projection. Discussion: The proposed model can be generalized to other unsupervised physiological signal learning tasks. Future directions include exploring task-specific data augmentations and combining self-supervised with supervised methods, building upon the initial success of self-supervised learning in this paper. Conclusions: We show that ContraWR is robust to noise and can provide high-quality EEG representations for downstream prediction tasks. In low-label scenarios (e.g., only 2% data has labels), ContraWR shows much better predictive power (e.g., 4% improvement on sleep staging accuracy) than supervised baselines.
SCRIB: Set-classifier with Class-specific Risk Bounds for Blackbox Models
Mar 05, 2021
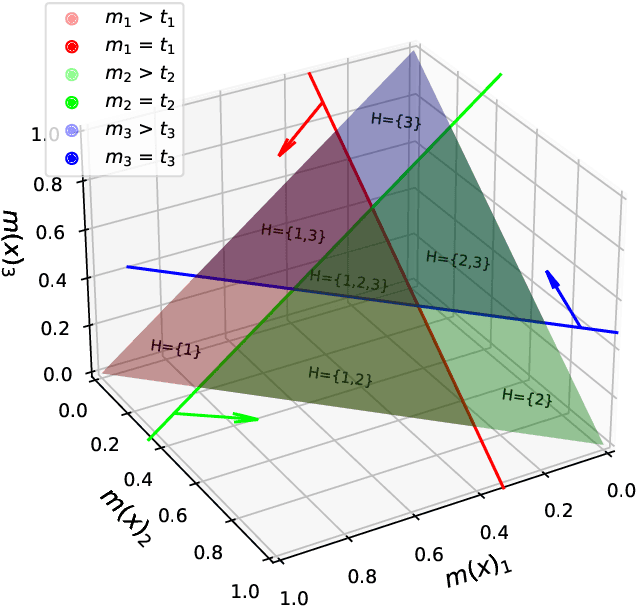
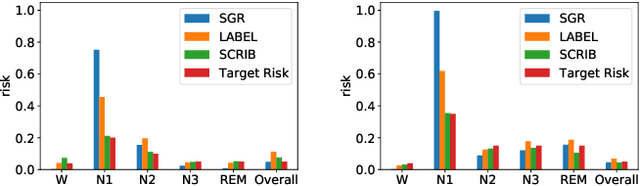

Abstract:Despite deep learning (DL) success in classification problems, DL classifiers do not provide a sound mechanism to decide when to refrain from predicting. Recent works tried to control the overall prediction risk with classification with rejection options. However, existing works overlook the different significance of different classes. We introduce Set-classifier with Class-specific RIsk Bounds (SCRIB) to tackle this problem, assigning multiple labels to each example. Given the output of a black-box model on the validation set, SCRIB constructs a set-classifier that controls the class-specific prediction risks with a theoretical guarantee. The key idea is to reject when the set classifier returns more than one label. We validated SCRIB on several medical applications, including sleep staging on electroencephalogram (EEG) data, X-ray COVID image classification, and atrial fibrillation detection based on electrocardiogram (ECG) data. SCRIB obtained desirable class-specific risks, which are 35\%-88\% closer to the target risks than baseline methods.
Sleep Apnea and Respiratory Anomaly Detection from a Wearable Band and Oxygen Saturation
Feb 24, 2021
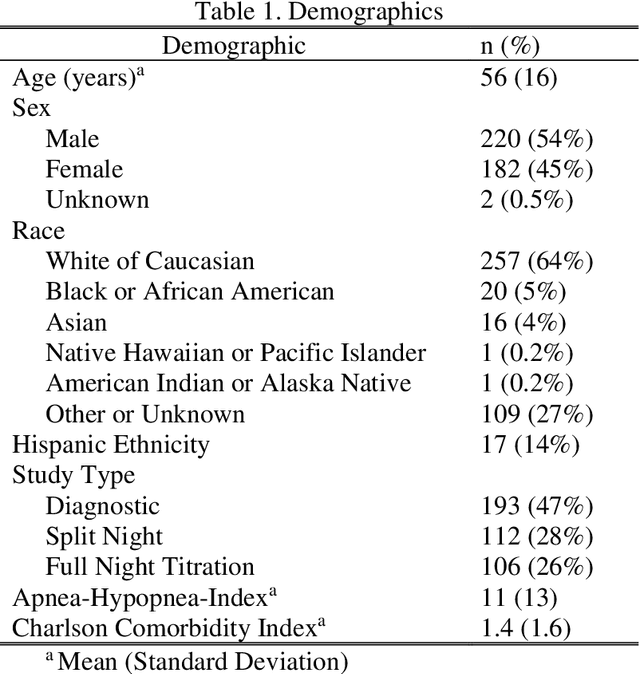
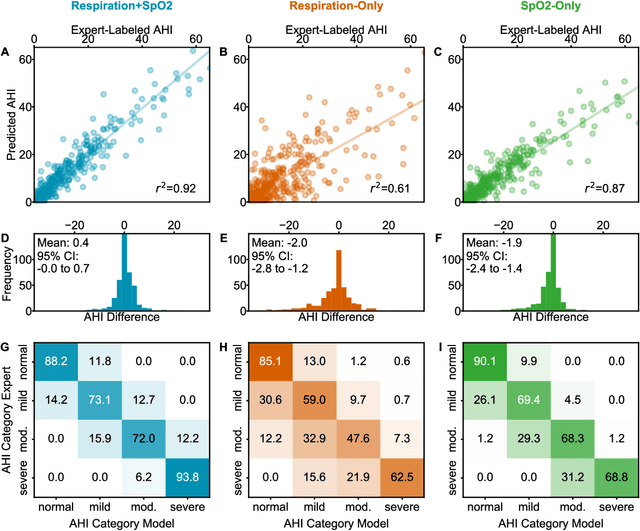
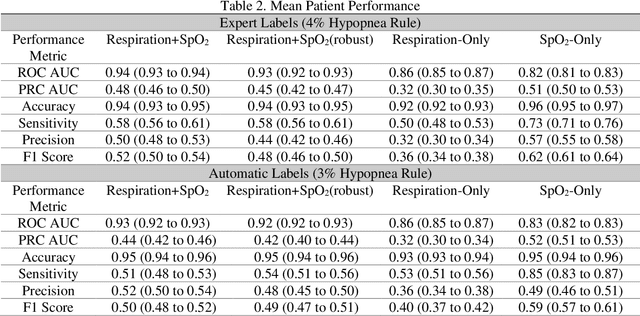
Abstract:Objective: Sleep related respiratory abnormalities are typically detected using polysomnography. There is a need in general medicine and critical care for a more convenient method to automatically detect sleep apnea from a simple, easy-to-wear device. The objective is to automatically detect abnormal respiration and estimate the Apnea-Hypopnea-Index (AHI) with a wearable respiratory device, compared to an SpO2 signal or polysomnography using a large (n = 412) dataset serving as ground truth. Methods: Simultaneously recorded polysomnographic (PSG) and wearable respiratory effort data were used to train and evaluate models in a cross-validation fashion. Time domain and complexity features were extracted, important features were identified, and a random forest model employed to detect events and predict AHI. Four models were trained: one each using the respiratory features only, a feature from the SpO2 (%)-signal only, and two additional models that use the respiratory features and the SpO2 (%)-feature, one allowing a time lag of 30 seconds between the two signals. Results: Event-based classification resulted in areas under the receiver operating characteristic curves of 0.94, 0.86, 0.82, and areas under the precision-recall curves of 0.48, 0.32, 0.51 for the models using respiration and SpO2, respiration-only, and SpO2-only respectively. Correlation between expert-labelled and predicted AHI was 0.96, 0.78, and 0.93, respectively. Conclusions: A wearable respiratory effort signal with or without SpO2 predicted AHI accurately. Given the large dataset and rigorous testing design, we expect our models are generalizable to evaluating respiration in a variety of environments, such as at home and in critical care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge