Wei-Ying Ma
S$^2$Drug: Bridging Protein Sequence and 3D Structure in Contrastive Representation Learning for Virtual Screening
Nov 10, 2025Abstract:Virtual screening (VS) is an essential task in drug discovery, focusing on the identification of small-molecule ligands that bind to specific protein pockets. Existing deep learning methods, from early regression models to recent contrastive learning approaches, primarily rely on structural data while overlooking protein sequences, which are more accessible and can enhance generalizability. However, directly integrating protein sequences poses challenges due to the redundancy and noise in large-scale protein-ligand datasets. To address these limitations, we propose \textbf{S$^2$Drug}, a two-stage framework that explicitly incorporates protein \textbf{S}equence information and 3D \textbf{S}tructure context in protein-ligand contrastive representation learning. In the first stage, we perform protein sequence pretraining on ChemBL using an ESM2-based backbone, combined with a tailored data sampling strategy to reduce redundancy and noise on both protein and ligand sides. In the second stage, we fine-tune on PDBBind by fusing sequence and structure information through a residue-level gating module, while introducing an auxiliary binding site prediction task. This auxiliary task guides the model to accurately localize binding residues within the protein sequence and capture their 3D spatial arrangement, thereby refining protein-ligand matching. Across multiple benchmarks, S$^2$Drug consistently improves virtual screening performance and achieves strong results on binding site prediction, demonstrating the value of bridging sequence and structure in contrastive learning.
FLEX: Continuous Agent Evolution via Forward Learning from Experience
Nov 09, 2025Abstract:Autonomous agents driven by Large Language Models (LLMs) have revolutionized reasoning and problem-solving but remain static after training, unable to grow with experience as intelligent beings do during deployment. We introduce Forward Learning with EXperience (FLEX), a gradient-free learning paradigm that enables LLM agents to continuously evolve through accumulated experience. Specifically, FLEX cultivates scalable and inheritable evolution by constructing a structured experience library through continual reflection on successes and failures during interaction with the environment. FLEX delivers substantial improvements on mathematical reasoning, chemical retrosynthesis, and protein fitness prediction (up to 23% on AIME25, 10% on USPTO50k, and 14% on ProteinGym). We further identify a clear scaling law of experiential growth and the phenomenon of experience inheritance across agents, marking a step toward scalable and inheritable continuous agent evolution. Project Page: https://flex-gensi-thuair.github.io.
Coder as Editor: Code-driven Interpretable Molecular Optimization
Oct 16, 2025Abstract:Molecular optimization is a central task in drug discovery that requires precise structural reasoning and domain knowledge. While large language models (LLMs) have shown promise in generating high-level editing intentions in natural language, they often struggle to faithfully execute these modifications-particularly when operating on non-intuitive representations like SMILES. We introduce MECo, a framework that bridges reasoning and execution by translating editing actions into executable code. MECo reformulates molecular optimization for LLMs as a cascaded framework: generating human-interpretable editing intentions from a molecule and property goal, followed by translating those intentions into executable structural edits via code generation. Our approach achieves over 98% accuracy in reproducing held-out realistic edits derived from chemical reactions and target-specific compound pairs. On downstream optimization benchmarks spanning physicochemical properties and target activities, MECo substantially improves consistency by 38-86 percentage points to 90%+ and achieves higher success rates over SMILES-based baselines while preserving structural similarity. By aligning intention with execution, MECo enables consistent, controllable and interpretable molecular design, laying the foundation for high-fidelity feedback loops and collaborative human-AI workflows in drug discovery.
ShortListing Model: A Streamlined SimplexDiffusion for Discrete Variable Generation
Aug 24, 2025Abstract:Generative modeling of discrete variables is challenging yet crucial for applications in natural language processing and biological sequence design. We introduce the Shortlisting Model (SLM), a novel simplex-based diffusion model inspired by progressive candidate pruning. SLM operates on simplex centroids, reducing generation complexity and enhancing scalability. Additionally, SLM incorporates a flexible implementation of classifier-free guidance, enhancing unconditional generation performance. Extensive experiments on DNA promoter and enhancer design, protein design, character-level and large-vocabulary language modeling demonstrate the competitive performance and strong potential of SLM. Our code can be found at https://github.com/GenSI-THUAIR/SLM
MemAgent: Reshaping Long-Context LLM with Multi-Conv RL-based Memory Agent
Jul 03, 2025

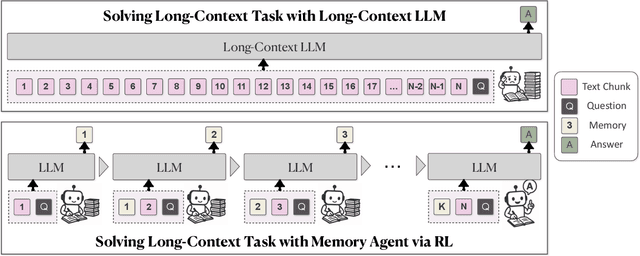
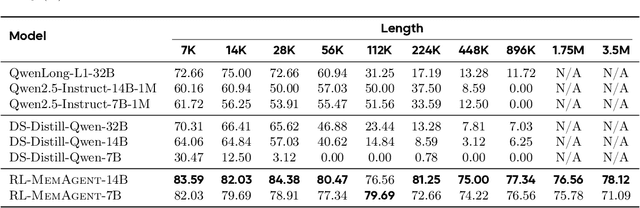
Abstract:Despite improvements by length extrapolation, efficient attention and memory modules, handling infinitely long documents with linear complexity without performance degradation during extrapolation remains the ultimate challenge in long-text processing. We directly optimize for long-text tasks in an end-to-end fashion and introduce a novel agent workflow, MemAgent, which reads text in segments and updates the memory using an overwrite strategy. We extend the DAPO algorithm to facilitate training via independent-context multi-conversation generation. MemAgent has demonstrated superb long-context capabilities, being able to extrapolate from an 8K context trained on 32K text to a 3.5M QA task with performance loss < 5% and achieves 95%+ in 512K RULER test.
AANet: Virtual Screening under Structural Uncertainty via Alignment and Aggregation
Jun 06, 2025Abstract:Virtual screening (VS) is a critical component of modern drug discovery, yet most existing methods--whether physics-based or deep learning-based--are developed around holo protein structures with known ligand-bound pockets. Consequently, their performance degrades significantly on apo or predicted structures such as those from AlphaFold2, which are more representative of real-world early-stage drug discovery, where pocket information is often missing. In this paper, we introduce an alignment-and-aggregation framework to enable accurate virtual screening under structural uncertainty. Our method comprises two core components: (1) a tri-modal contrastive learning module that aligns representations of the ligand, the holo pocket, and cavities detected from structures, thereby enhancing robustness to pocket localization error; and (2) a cross-attention based adapter for dynamically aggregating candidate binding sites, enabling the model to learn from activity data even without precise pocket annotations. We evaluated our method on a newly curated benchmark of apo structures, where it significantly outperforms state-of-the-art methods in blind apo setting, improving the early enrichment factor (EF1%) from 11.75 to 37.19. Notably, it also maintains strong performance on holo structures. These results demonstrate the promise of our approach in advancing first-in-class drug discovery, particularly in scenarios lacking experimentally resolved protein-ligand complexes.
Piloting Structure-Based Drug Design via Modality-Specific Optimal Schedule
May 12, 2025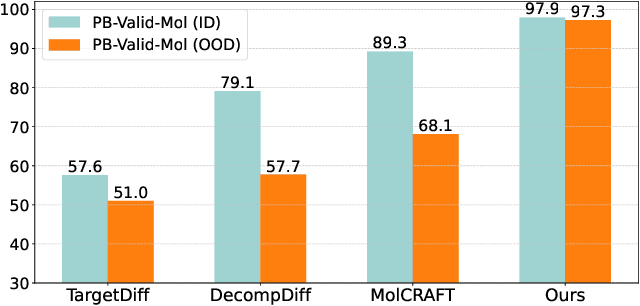
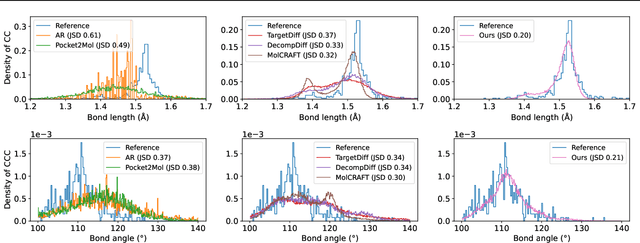
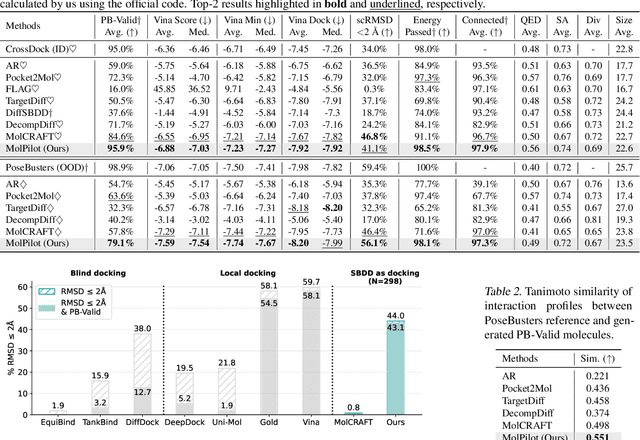
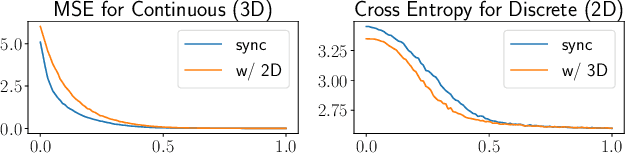
Abstract:Structure-Based Drug Design (SBDD) is crucial for identifying bioactive molecules. Recent deep generative models are faced with challenges in geometric structure modeling. A major bottleneck lies in the twisted probability path of multi-modalities -- continuous 3D positions and discrete 2D topologies -- which jointly determine molecular geometries. By establishing the fact that noise schedules decide the Variational Lower Bound (VLB) for the twisted probability path, we propose VLB-Optimal Scheduling (VOS) strategy in this under-explored area, which optimizes VLB as a path integral for SBDD. Our model effectively enhances molecular geometries and interaction modeling, achieving state-of-the-art PoseBusters passing rate of 95.9% on CrossDock, more than 10% improvement upon strong baselines, while maintaining high affinities and robust intramolecular validity evaluated on held-out test set.
PharmAgents: Building a Virtual Pharma with Large Language Model Agents
Mar 31, 2025Abstract:The discovery of novel small molecule drugs remains a critical scientific challenge with far-reaching implications for treating diseases and advancing human health. Traditional drug development--especially for small molecule therapeutics--is a highly complex, resource-intensive, and time-consuming process that requires multidisciplinary collaboration. Recent breakthroughs in artificial intelligence (AI), particularly the rise of large language models (LLMs), present a transformative opportunity to streamline and accelerate this process. In this paper, we introduce PharmAgents, a virtual pharmaceutical ecosystem driven by LLM-based multi-agent collaboration. PharmAgents simulates the full drug discovery workflow--from target discovery to preclinical evaluation--by integrating explainable, LLM-driven agents equipped with specialized machine learning models and computational tools. Through structured knowledge exchange and automated optimization, PharmAgents identifies potential therapeutic targets, discovers promising lead compounds, enhances binding affinity and key molecular properties, and performs in silico analyses of toxicity and synthetic feasibility. Additionally, the system supports interpretability, agent interaction, and self-evolvement, enabling it to refine future drug designs based on prior experience. By showcasing the potential of LLM-powered multi-agent systems in drug discovery, this work establishes a new paradigm for autonomous, explainable, and scalable pharmaceutical research, with future extensions toward comprehensive drug lifecycle management.
DAPO: An Open-Source LLM Reinforcement Learning System at Scale
Mar 18, 2025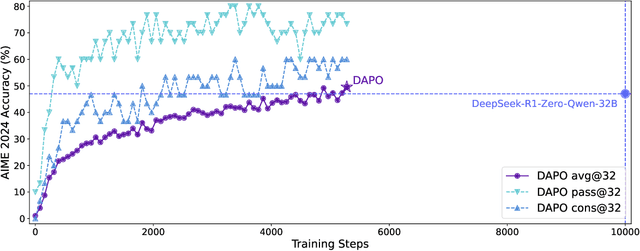
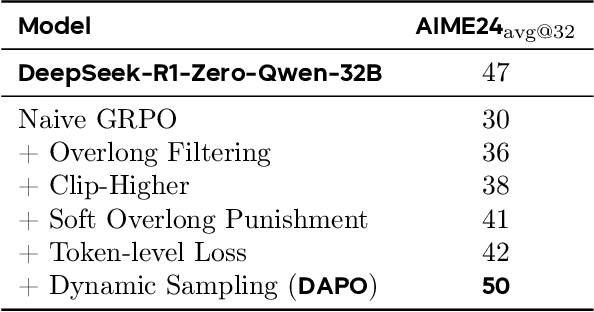
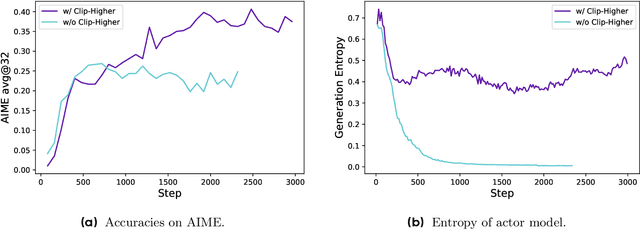

Abstract:Inference scaling empowers LLMs with unprecedented reasoning ability, with reinforcement learning as the core technique to elicit complex reasoning. However, key technical details of state-of-the-art reasoning LLMs are concealed (such as in OpenAI o1 blog and DeepSeek R1 technical report), thus the community still struggles to reproduce their RL training results. We propose the $\textbf{D}$ecoupled Clip and $\textbf{D}$ynamic s$\textbf{A}$mpling $\textbf{P}$olicy $\textbf{O}$ptimization ($\textbf{DAPO}$) algorithm, and fully open-source a state-of-the-art large-scale RL system that achieves 50 points on AIME 2024 using Qwen2.5-32B base model. Unlike previous works that withhold training details, we introduce four key techniques of our algorithm that make large-scale LLM RL a success. In addition, we open-source our training code, which is built on the verl framework, along with a carefully curated and processed dataset. These components of our open-source system enhance reproducibility and support future research in large-scale LLM RL.
Straight-Line Diffusion Model for Efficient 3D Molecular Generation
Mar 04, 2025Abstract:Diffusion-based models have shown great promise in molecular generation but often require a large number of sampling steps to generate valid samples. In this paper, we introduce a novel Straight-Line Diffusion Model (SLDM) to tackle this problem, by formulating the diffusion process to follow a linear trajectory. The proposed process aligns well with the noise sensitivity characteristic of molecular structures and uniformly distributes reconstruction effort across the generative process, thus enhancing learning efficiency and efficacy. Consequently, SLDM achieves state-of-the-art performance on 3D molecule generation benchmarks, delivering a 100-fold improvement in sampling efficiency. Furthermore, experiments on toy data and image generation tasks validate the generality and robustness of SLDM, showcasing its potential across diverse generative modeling domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge