Haichuan Tan
AANet: Virtual Screening under Structural Uncertainty via Alignment and Aggregation
Jun 06, 2025Abstract:Virtual screening (VS) is a critical component of modern drug discovery, yet most existing methods--whether physics-based or deep learning-based--are developed around holo protein structures with known ligand-bound pockets. Consequently, their performance degrades significantly on apo or predicted structures such as those from AlphaFold2, which are more representative of real-world early-stage drug discovery, where pocket information is often missing. In this paper, we introduce an alignment-and-aggregation framework to enable accurate virtual screening under structural uncertainty. Our method comprises two core components: (1) a tri-modal contrastive learning module that aligns representations of the ligand, the holo pocket, and cavities detected from structures, thereby enhancing robustness to pocket localization error; and (2) a cross-attention based adapter for dynamically aggregating candidate binding sites, enabling the model to learn from activity data even without precise pocket annotations. We evaluated our method on a newly curated benchmark of apo structures, where it significantly outperforms state-of-the-art methods in blind apo setting, improving the early enrichment factor (EF1%) from 11.75 to 37.19. Notably, it also maintains strong performance on holo structures. These results demonstrate the promise of our approach in advancing first-in-class drug discovery, particularly in scenarios lacking experimentally resolved protein-ligand complexes.
From Theory to Therapy: Reframing SBDD Model Evaluation via Practical Metrics
Jun 13, 2024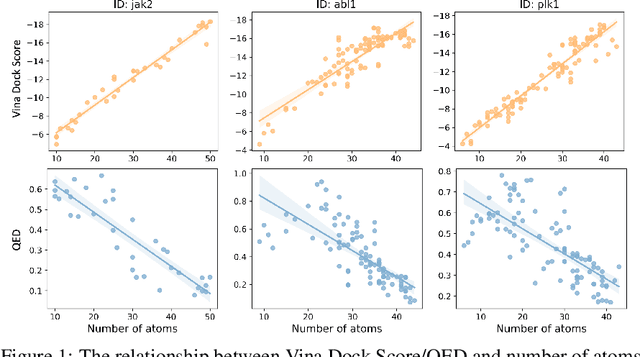
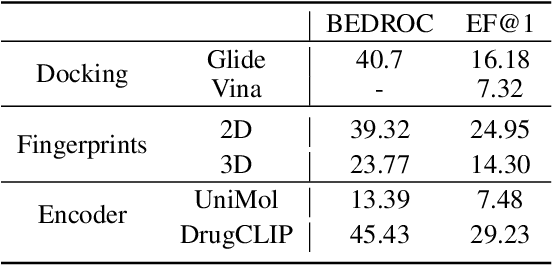
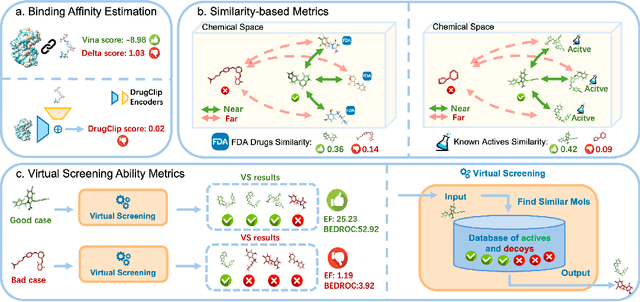
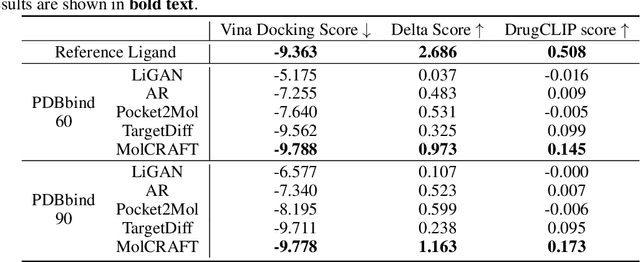
Abstract:Recent advancements in structure-based drug design (SBDD) have significantly enhanced the efficiency and precision of drug discovery by generating molecules tailored to bind specific protein pockets. Despite these technological strides, their practical application in real-world drug development remains challenging due to the complexities of synthesizing and testing these molecules. The reliability of the Vina docking score, the current standard for assessing binding abilities, is increasingly questioned due to its susceptibility to overfitting. To address these limitations, we propose a comprehensive evaluation framework that includes assessing the similarity of generated molecules to known active compounds, introducing a virtual screening-based metric for practical deployment capabilities, and re-evaluating binding affinity more rigorously. Our experiments reveal that while current SBDD models achieve high Vina scores, they fall short in practical usability metrics, highlighting a significant gap between theoretical predictions and real-world applicability. Our proposed metrics and dataset aim to bridge this gap, enhancing the practical applicability of future SBDD models and aligning them more closely with the needs of pharmaceutical research and development.
DrugCLIP: Contrastive Protein-Molecule Representation Learning for Virtual Screening
Oct 10, 2023Abstract:Virtual screening, which identifies potential drugs from vast compound databases to bind with a particular protein pocket, is a critical step in AI-assisted drug discovery. Traditional docking methods are highly time-consuming, and can only work with a restricted search library in real-life applications. Recent supervised learning approaches using scoring functions for binding-affinity prediction, although promising, have not yet surpassed docking methods due to their strong dependency on limited data with reliable binding-affinity labels. In this paper, we propose a novel contrastive learning framework, DrugCLIP, by reformulating virtual screening as a dense retrieval task and employing contrastive learning to align representations of binding protein pockets and molecules from a large quantity of pairwise data without explicit binding-affinity scores. We also introduce a biological-knowledge inspired data augmentation strategy to learn better protein-molecule representations. Extensive experiments show that DrugCLIP significantly outperforms traditional docking and supervised learning methods on diverse virtual screening benchmarks with highly reduced computation time, especially in zero-shot setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge