Yuxuan Song
Stable-DiffCoder: Pushing the Frontier of Code Diffusion Large Language Model
Jan 23, 2026Abstract:Diffusion-based language models (DLLMs) offer non-sequential, block-wise generation and richer data reuse compared to autoregressive (AR) models, but existing code DLLMs still lag behind strong AR baselines under comparable budgets. We revisit this setting in a controlled study and introduce Stable-DiffCoder, a block diffusion code model that reuses the Seed-Coder architecture, data, and training pipeline. To enable efficient knowledge learning and stable training, we incorporate a block diffusion continual pretraining (CPT) stage enhanced by a tailored warmup and block-wise clipped noise schedule. Under the same data and architecture, Stable-DiffCoder overall outperforms its AR counterpart on a broad suite of code benchmarks. Moreover, relying only on the CPT and supervised fine-tuning stages, Stable-DiffCoder achieves stronger performance than a wide range of \~8B ARs and DLLMs, demonstrating that diffusion-based training can improve code modeling quality beyond AR training alone. Moreover, diffusion-based any-order modeling improves structured code modeling for editing and reasoning, and through data augmentation, benefits low-resource coding languages.
Dynamic Large Concept Models: Latent Reasoning in an Adaptive Semantic Space
Dec 31, 2025Abstract:Large Language Models (LLMs) apply uniform computation to all tokens, despite language exhibiting highly non-uniform information density. This token-uniform regime wastes capacity on locally predictable spans while under-allocating computation to semantically critical transitions. We propose $\textbf{Dynamic Large Concept Models (DLCM)}$, a hierarchical language modeling framework that learns semantic boundaries from latent representations and shifts computation from tokens to a compressed concept space where reasoning is more efficient. DLCM discovers variable-length concepts end-to-end without relying on predefined linguistic units. Hierarchical compression fundamentally changes scaling behavior. We introduce the first $\textbf{compression-aware scaling law}$, which disentangles token-level capacity, concept-level reasoning capacity, and compression ratio, enabling principled compute allocation under fixed FLOPs. To stably train this heterogeneous architecture, we further develop a $\textbf{decoupled $μ$P parametrization}$ that supports zero-shot hyperparameter transfer across widths and compression regimes. At a practical setting ($R=4$, corresponding to an average of four tokens per concept), DLCM reallocates roughly one-third of inference compute into a higher-capacity reasoning backbone, achieving a $\textbf{+2.69$\%$ average improvement}$ across 12 zero-shot benchmarks under matched inference FLOPs.
ShortListing Model: A Streamlined SimplexDiffusion for Discrete Variable Generation
Aug 24, 2025Abstract:Generative modeling of discrete variables is challenging yet crucial for applications in natural language processing and biological sequence design. We introduce the Shortlisting Model (SLM), a novel simplex-based diffusion model inspired by progressive candidate pruning. SLM operates on simplex centroids, reducing generation complexity and enhancing scalability. Additionally, SLM incorporates a flexible implementation of classifier-free guidance, enhancing unconditional generation performance. Extensive experiments on DNA promoter and enhancer design, protein design, character-level and large-vocabulary language modeling demonstrate the competitive performance and strong potential of SLM. Our code can be found at https://github.com/GenSI-THUAIR/SLM
Protenix-Mini: Efficient Structure Predictor via Compact Architecture, Few-Step Diffusion and Switchable pLM
Jul 16, 2025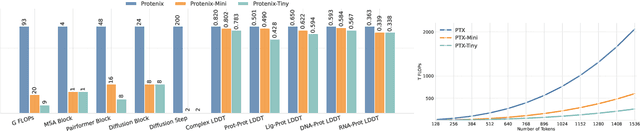
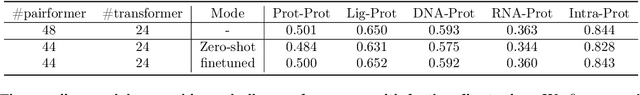
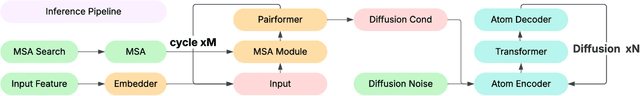
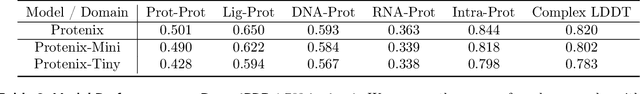
Abstract:Lightweight inference is critical for biomolecular structure prediction and other downstream tasks, enabling efficient real-world deployment and inference-time scaling for large-scale applications. In this work, we address the challenge of balancing model efficiency and prediction accuracy by making several key modifications, 1) Multi-step AF3 sampler is replaced by a few-step ODE sampler, significantly reducing computational overhead for the diffusion module part during inference; 2) In the open-source Protenix framework, a subset of pairformer or diffusion transformer blocks doesn't make contributions to the final structure prediction, presenting opportunities for architectural pruning and lightweight redesign; 3) A model incorporating an ESM module is trained to substitute the conventional MSA module, reducing MSA preprocessing time. Building on these key insights, we present Protenix-Mini, a compact and optimized model designed for efficient protein structure prediction. This streamlined version incorporates a more efficient architectural design with a two-step Ordinary Differential Equation (ODE) sampling strategy. By eliminating redundant Transformer components and refining the sampling process, Protenix-Mini significantly reduces model complexity with slight accuracy drop. Evaluations on benchmark datasets demonstrate that it achieves high-fidelity predictions, with only a negligible 1 to 5 percent decrease in performance on benchmark datasets compared to its full-scale counterpart. This makes Protenix-Mini an ideal choice for applications where computational resources are limited but accurate structure prediction remains crucial.
Piloting Structure-Based Drug Design via Modality-Specific Optimal Schedule
May 12, 2025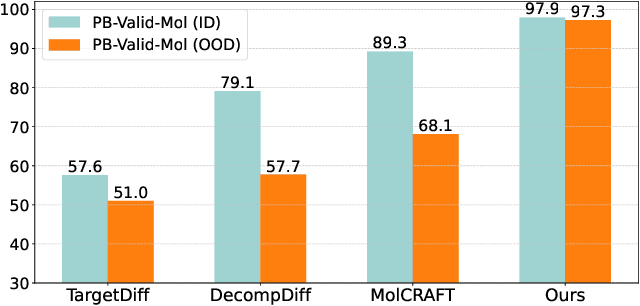
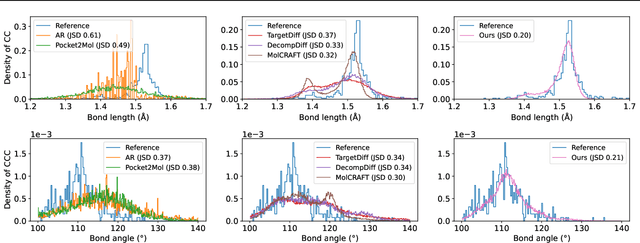
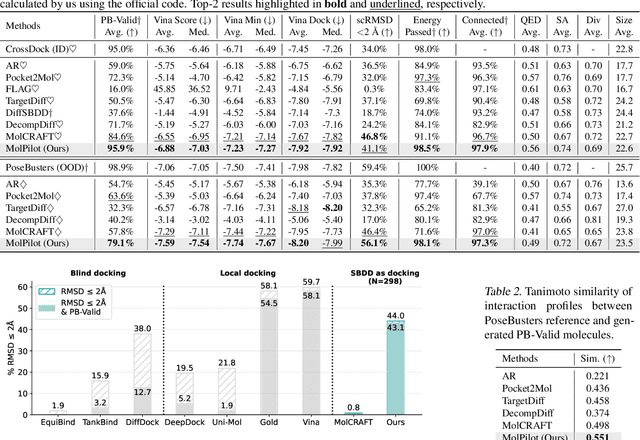
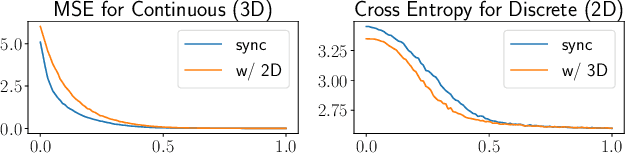
Abstract:Structure-Based Drug Design (SBDD) is crucial for identifying bioactive molecules. Recent deep generative models are faced with challenges in geometric structure modeling. A major bottleneck lies in the twisted probability path of multi-modalities -- continuous 3D positions and discrete 2D topologies -- which jointly determine molecular geometries. By establishing the fact that noise schedules decide the Variational Lower Bound (VLB) for the twisted probability path, we propose VLB-Optimal Scheduling (VOS) strategy in this under-explored area, which optimizes VLB as a path integral for SBDD. Our model effectively enhances molecular geometries and interaction modeling, achieving state-of-the-art PoseBusters passing rate of 95.9% on CrossDock, more than 10% improvement upon strong baselines, while maintaining high affinities and robust intramolecular validity evaluated on held-out test set.
DAPO: An Open-Source LLM Reinforcement Learning System at Scale
Mar 18, 2025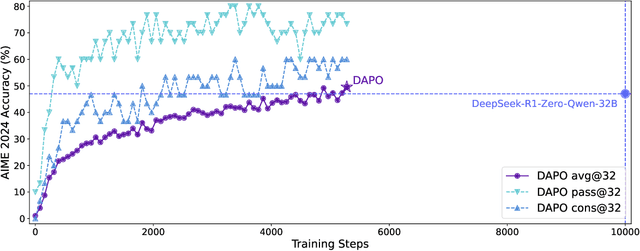
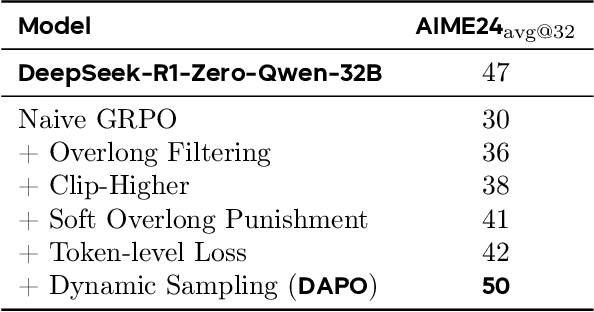
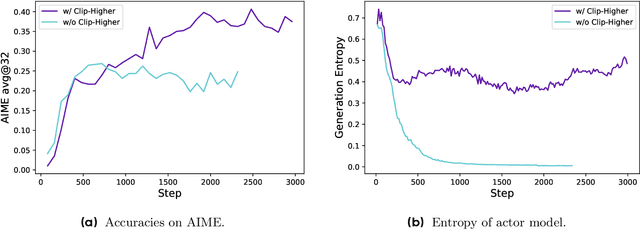

Abstract:Inference scaling empowers LLMs with unprecedented reasoning ability, with reinforcement learning as the core technique to elicit complex reasoning. However, key technical details of state-of-the-art reasoning LLMs are concealed (such as in OpenAI o1 blog and DeepSeek R1 technical report), thus the community still struggles to reproduce their RL training results. We propose the $\textbf{D}$ecoupled Clip and $\textbf{D}$ynamic s$\textbf{A}$mpling $\textbf{P}$olicy $\textbf{O}$ptimization ($\textbf{DAPO}$) algorithm, and fully open-source a state-of-the-art large-scale RL system that achieves 50 points on AIME 2024 using Qwen2.5-32B base model. Unlike previous works that withhold training details, we introduce four key techniques of our algorithm that make large-scale LLM RL a success. In addition, we open-source our training code, which is built on the verl framework, along with a carefully curated and processed dataset. These components of our open-source system enhance reproducibility and support future research in large-scale LLM RL.
A Periodic Bayesian Flow for Material Generation
Feb 04, 2025



Abstract:Generative modeling of crystal data distribution is an important yet challenging task due to the unique periodic physical symmetry of crystals. Diffusion-based methods have shown early promise in modeling crystal distribution. More recently, Bayesian Flow Networks were introduced to aggregate noisy latent variables, resulting in a variance-reduced parameter space that has been shown to be advantageous for modeling Euclidean data distributions with structural constraints (Song et al., 2023). Inspired by this, we seek to unlock its potential for modeling variables located in non-Euclidean manifolds e.g. those within crystal structures, by overcoming challenging theoretical issues. We introduce CrysBFN, a novel crystal generation method by proposing a periodic Bayesian flow, which essentially differs from the original Gaussian-based BFN by exhibiting non-monotonic entropy dynamics. To successfully realize the concept of periodic Bayesian flow, CrysBFN integrates a new entropy conditioning mechanism and empirically demonstrates its significance compared to time-conditioning. Extensive experiments over both crystal ab initio generation and crystal structure prediction tasks demonstrate the superiority of CrysBFN, which consistently achieves new state-of-the-art on all benchmarks. Surprisingly, we found that CrysBFN enjoys a significant improvement in sampling efficiency, e.g., ~100x speedup 10 v.s. 2000 steps network forwards) compared with previous diffusion-based methods on MP-20 dataset. Code is available at https://github.com/wu-han-lin/CrysBFN.
ACDiT: Interpolating Autoregressive Conditional Modeling and Diffusion Transformer
Dec 10, 2024Abstract:The recent surge of interest in comprehensive multimodal models has necessitated the unification of diverse modalities. However, the unification suffers from disparate methodologies. Continuous visual generation necessitates the full-sequence diffusion-based approach, despite its divergence from the autoregressive modeling in the text domain. We posit that autoregressive modeling, i.e., predicting the future based on past deterministic experience, remains crucial in developing both a visual generation model and a potential unified multimodal model. In this paper, we explore an interpolation between the autoregressive modeling and full-parameters diffusion to model visual information. At its core, we present ACDiT, an Autoregressive blockwise Conditional Diffusion Transformer, where the block size of diffusion, i.e., the size of autoregressive units, can be flexibly adjusted to interpolate between token-wise autoregression and full-sequence diffusion. ACDiT is easy to implement, as simple as creating a Skip-Causal Attention Mask (SCAM) during training. During inference, the process iterates between diffusion denoising and autoregressive decoding that can make full use of KV-Cache. We verify the effectiveness of ACDiT on image and video generation tasks. We also demonstrate that benefitted from autoregressive modeling, ACDiT can be seamlessly used in visual understanding tasks despite being trained on the diffusion objective. The analysis of the trade-off between autoregressive modeling and diffusion demonstrates the potential of ACDiT to be used in long-horizon visual generation tasks. These strengths make it promising as the backbone of future unified models.
Structure-Based Molecule Optimization via Gradient-Guided Bayesian Update
Nov 21, 2024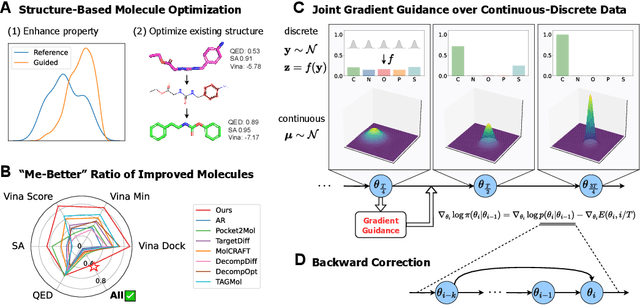
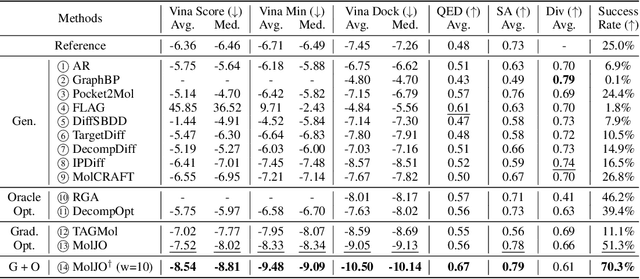
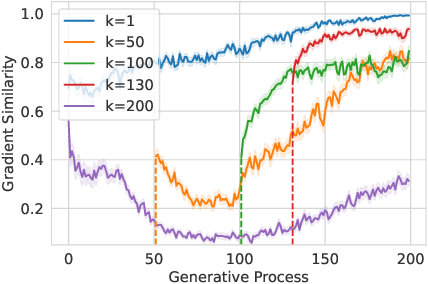
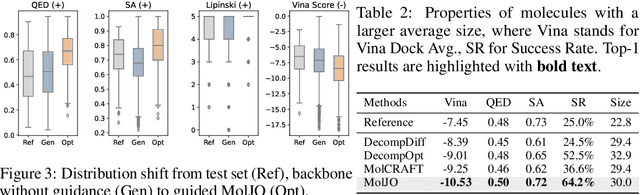
Abstract:Structure-based molecule optimization (SBMO) aims to optimize molecules with both continuous coordinates and discrete types against protein targets. A promising direction is to exert gradient guidance on generative models given its remarkable success in images, but it is challenging to guide discrete data and risks inconsistencies between modalities. To this end, we leverage a continuous and differentiable space derived through Bayesian inference, presenting Molecule Joint Optimization (MolJO), the first gradient-based SBMO framework that facilitates joint guidance signals across different modalities while preserving SE(3)-equivariance. We introduce a novel backward correction strategy that optimizes within a sliding window of the past histories, allowing for a seamless trade-off between explore-and-exploit during optimization. Our proposed MolJO achieves state-of-the-art performance on CrossDocked2020 benchmark (Success Rate 51.3% , Vina Dock -9.05 and SA 0.78), more than 4x improvement in Success Rate compared to the gradient-based counterpart, and 2x "Me-Better" Ratio as much as 3D baselines. Furthermore, we extend MolJO to a wide range of optimization settings, including multi-objective optimization and challenging tasks in drug design such as R-group optimization and scaffold hopping, further underscoring its versatility and potential.
$f$-PO: Generalizing Preference Optimization with $f$-divergence Minimization
Oct 29, 2024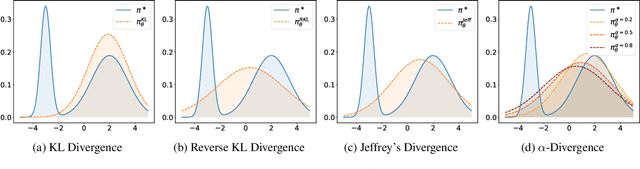
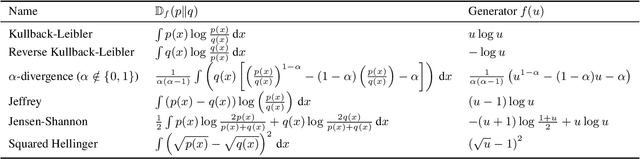
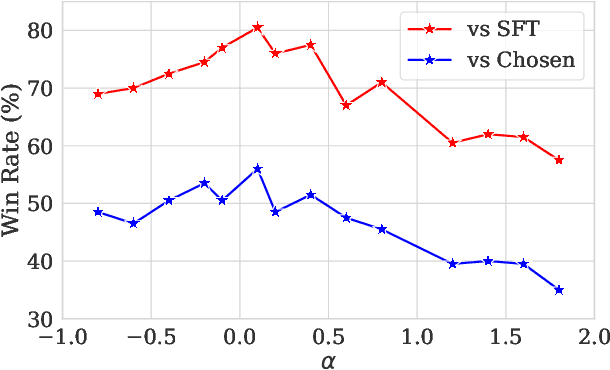

Abstract:Preference optimization has made significant progress recently, with numerous methods developed to align language models with human preferences. This paper introduces $f$-divergence Preference Optimization ($f$-PO), a novel framework that generalizes and extends existing approaches. $f$-PO minimizes $f$-divergences between the optimized policy and the optimal policy, encompassing a broad family of alignment methods using various divergences. Our approach unifies previous algorithms like DPO and EXO, while offering new variants through different choices of $f$-divergences. We provide theoretical analysis of $f$-PO's properties and conduct extensive experiments on state-of-the-art language models using benchmark datasets. Results demonstrate $f$-PO's effectiveness across various tasks, achieving superior performance compared to existing methods on popular benchmarks such as AlpacaEval 2, Arena-Hard, and MT-Bench. Additionally, we present ablation studies exploring the impact of different $f$-divergences, offering insights into the trade-offs between regularization and performance in offline preference optimization. Our work contributes both practical algorithms and theoretical understanding to the field of language model alignment. Code is available at https://github.com/MinkaiXu/fPO.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge