Songchi Zhou
Baichuan-M1: Pushing the Medical Capability of Large Language Models
Feb 18, 2025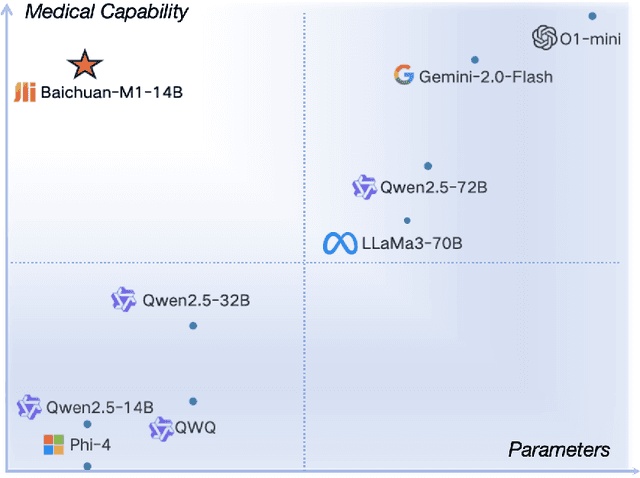
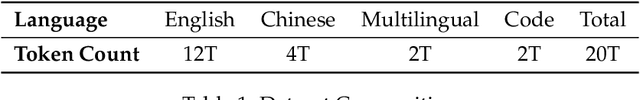
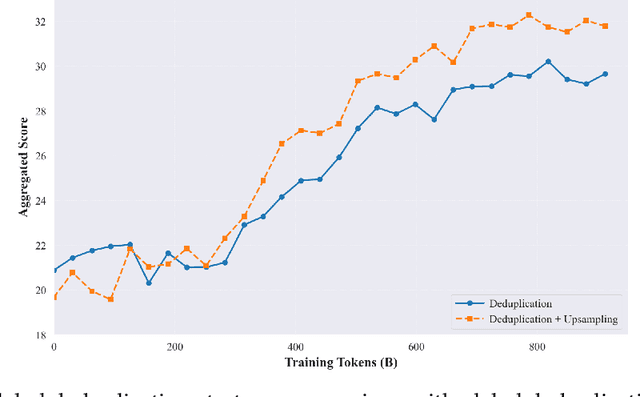
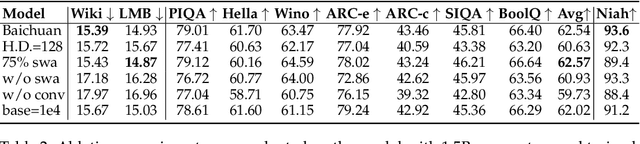
Abstract:The current generation of large language models (LLMs) is typically designed for broad, general-purpose applications, while domain-specific LLMs, especially in vertical fields like medicine, remain relatively scarce. In particular, the development of highly efficient and practical LLMs for the medical domain is challenging due to the complexity of medical knowledge and the limited availability of high-quality data. To bridge this gap, we introduce Baichuan-M1, a series of large language models specifically optimized for medical applications. Unlike traditional approaches that simply continue pretraining on existing models or apply post-training to a general base model, Baichuan-M1 is trained from scratch with a dedicated focus on enhancing medical capabilities. Our model is trained on 20 trillion tokens and incorporates a range of effective training methods that strike a balance between general capabilities and medical expertise. As a result, Baichuan-M1 not only performs strongly across general domains such as mathematics and coding but also excels in specialized medical fields. We have open-sourced Baichuan-M1-14B, a mini version of our model, which can be accessed through the following links.
Evaluating Entity Retrieval in Electronic Health Records: a Semantic Gap Perspective
Feb 10, 2025



Abstract:Entity retrieval plays a crucial role in the utilization of Electronic Health Records (EHRs) and is applied across a wide range of clinical practices. However, a comprehensive evaluation of this task is lacking due to the absence of a public benchmark. In this paper, we propose the development and release of a novel benchmark for evaluating entity retrieval in EHRs, with a particular focus on the semantic gap issue. Using discharge summaries from the MIMIC-III dataset, we incorporate ICD codes and prescription labels associated with the notes as queries, and annotate relevance judgments using GPT-4. In total, we use 1,000 patient notes, generate 1,246 queries, and provide over 77,000 relevance annotations. To offer the first assessment of the semantic gap, we introduce a novel classification system for relevance matches. Leveraging GPT-4, we categorize each relevant pair into one of five categories: string, synonym, abbreviation, hyponym, and implication. Using the proposed benchmark, we evaluate several retrieval methods, including BM25, query expansion, and state-of-the-art dense retrievers. Our findings show that BM25 provides a strong baseline but struggles with semantic matches. Query expansion significantly improves performance, though it slightly reduces string match capabilities. Dense retrievers outperform traditional methods, particularly for semantic matches, and general-domain dense retrievers often surpass those trained specifically in the biomedical domain.
Annotation of Sleep Depth Index with Scalable Deep Learning Yields Novel Digital Biomarkers for Sleep Health
Jul 05, 2024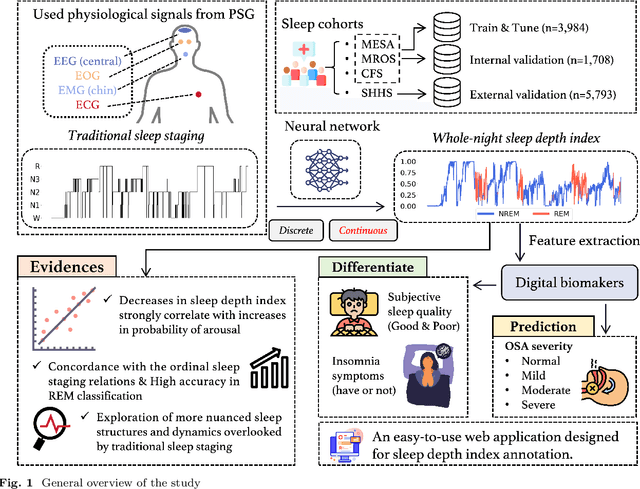

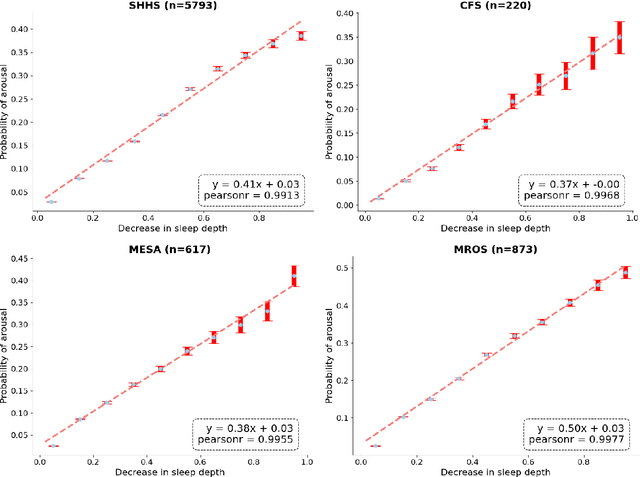
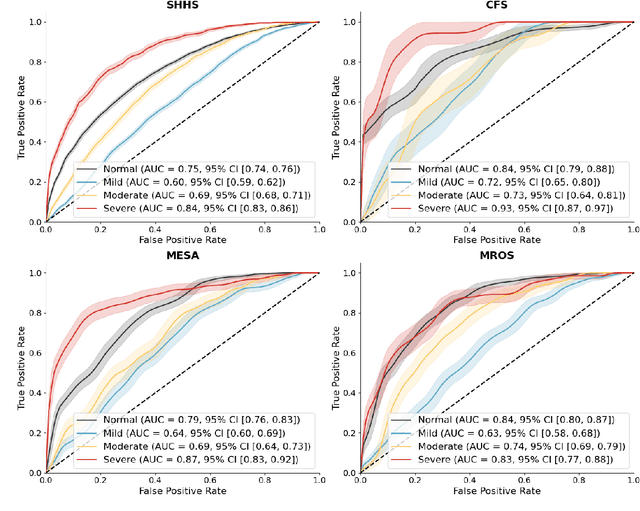
Abstract:Traditional sleep staging categorizes sleep and wakefulness into five coarse-grained classes, overlooking subtle variations within each stage. It provides limited information about the probability of arousal and may hinder the diagnosis of sleep disorders, such as insomnia. To address this issue, we propose a deep-learning method for automatic and scalable annotation of sleep depth index using existing sleep staging labels. Our approach is validated using polysomnography from over ten thousand recordings across four large-scale cohorts. The results show a strong correlation between the decrease in sleep depth index and the increase in arousal likelihood. Several case studies indicate that the sleep depth index captures more nuanced sleep structures than conventional sleep staging. Sleep biomarkers extracted from the whole-night sleep depth index exhibit statistically significant differences with medium-to-large effect sizes across groups of varied subjective sleep quality and insomnia symptoms. These sleep biomarkers also promise utility in predicting the severity of obstructive sleep apnea, particularly in severe cases. Our study underscores the utility of the proposed method for continuous sleep depth annotation, which could reveal more detailed structures and dynamics within whole-night sleep and yield novel digital biomarkers beneficial for sleep health.
High-throughput Biomedical Relation Extraction for Semi-Structured Web Articles Empowered by Large Language Models
Dec 15, 2023Abstract:Objective: To develop a high-throughput biomedical relation extraction system that takes advantage of the large language models' (LLMs) reading comprehension ability and biomedical world knowledge in a scalable and evidential manner. Methods: We formulate the relation extraction task as a simple binary classification problem for large language models such as ChatGPT. Specifically, LLMs make the decision based on the external corpus and its world knowledge, giving the reason for the judgment to factual verification. This method is tailored for semi-structured web articles, wherein we designate the main title as the tail entity and explicitly incorporate it into the context, and the potential head entities are matched based on a biomedical thesaurus. Moreover, lengthy contents are sliced into text chunks, embedded, and retrieved with additional embedding models, ensuring compatibility with the context window size constraints of available open-source LLMs. Results: Using an open-source LLM, we extracted 304315 relation triplets of three distinct relation types from four reputable biomedical websites. To assess the efficacy of the basic pipeline employed for biomedical relation extraction, we curated a benchmark dataset annotated by a medical expert. Evaluation results indicate that the pipeline exhibits performance comparable to that of GPT-4. Case studies further illuminate challenges faced by contemporary LLMs in the context of biomedical relation extraction for semi-structured web articles. Conclusion: The proposed method has demonstrated its effectiveness in leveraging the strengths of LLMs for high-throughput biomedical relation extraction. Its adaptability is evident, as it can be seamlessly extended to diverse semi-structured biomedical websites, facilitating the extraction of various types of biomedical relations with ease.
EHRDiff: Exploring Realistic EHR Synthesis with Diffusion Models
Mar 10, 2023



Abstract:Electronic health records (EHR) contain vast biomedical knowledge and are rich resources for developing precise medicine systems. However, due to privacy concerns, there are limited high-quality EHR data accessible to researchers hence hindering the advancement of methodologies. Recent research has explored using generative modelling methods to synthesize realistic EHR data, and most proposed methods are based on the generative adversarial network (GAN) and its variants for EHR synthesis. Although GAN-style methods achieved state-of-the-art performance in generating high-quality EHR data, such methods are hard to train and prone to mode collapse. Diffusion models are recently proposed generative modelling methods and set cutting-edge performance in image generation. The performance of diffusion models in realistic EHR synthesis is rarely explored. In this work, we explore whether the superior performance of diffusion models can translate to the domain of EHR synthesis and propose a novel EHR synthesis method named EHRDiff. Through comprehensive experiments, EHRDiff achieves new state-of-the-art performance for the quality of synthetic EHR data and can better protect private information in real training EHRs in the meanwhile.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge