Zhengyun Zhao
DR.EHR: Dense Retrieval for Electronic Health Record with Knowledge Injection and Synthetic Data
Jul 24, 2025Abstract:Electronic Health Records (EHRs) are pivotal in clinical practices, yet their retrieval remains a challenge mainly due to semantic gap issues. Recent advancements in dense retrieval offer promising solutions but existing models, both general-domain and biomedical-domain, fall short due to insufficient medical knowledge or mismatched training corpora. This paper introduces \texttt{DR.EHR}, a series of dense retrieval models specifically tailored for EHR retrieval. We propose a two-stage training pipeline utilizing MIMIC-IV discharge summaries to address the need for extensive medical knowledge and large-scale training data. The first stage involves medical entity extraction and knowledge injection from a biomedical knowledge graph, while the second stage employs large language models to generate diverse training data. We train two variants of \texttt{DR.EHR}, with 110M and 7B parameters, respectively. Evaluated on the CliniQ benchmark, our models significantly outperforms all existing dense retrievers, achieving state-of-the-art results. Detailed analyses confirm our models' superiority across various match and query types, particularly in challenging semantic matches like implication and abbreviation. Ablation studies validate the effectiveness of each pipeline component, and supplementary experiments on EHR QA datasets demonstrate the models' generalizability on natural language questions, including complex ones with multiple entities. This work significantly advances EHR retrieval, offering a robust solution for clinical applications.
Baichuan-M1: Pushing the Medical Capability of Large Language Models
Feb 18, 2025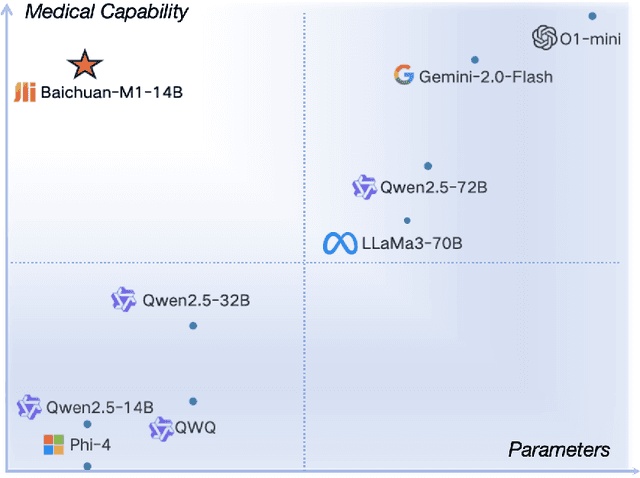
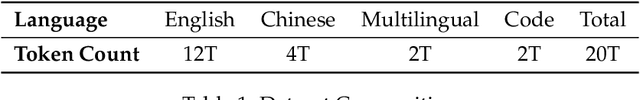
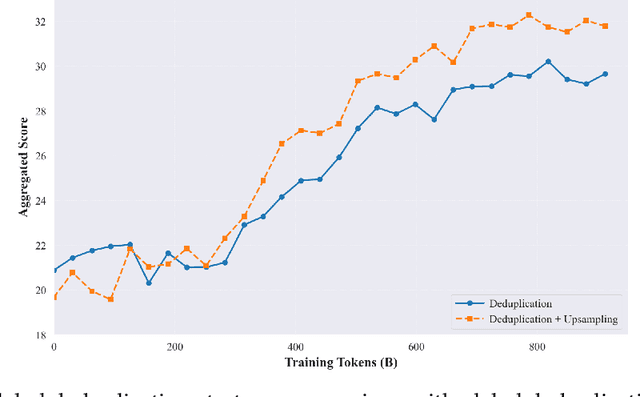
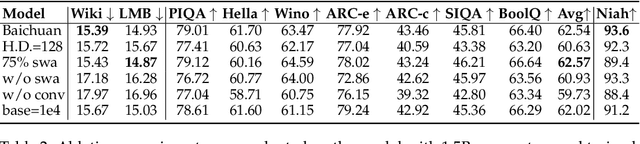
Abstract:The current generation of large language models (LLMs) is typically designed for broad, general-purpose applications, while domain-specific LLMs, especially in vertical fields like medicine, remain relatively scarce. In particular, the development of highly efficient and practical LLMs for the medical domain is challenging due to the complexity of medical knowledge and the limited availability of high-quality data. To bridge this gap, we introduce Baichuan-M1, a series of large language models specifically optimized for medical applications. Unlike traditional approaches that simply continue pretraining on existing models or apply post-training to a general base model, Baichuan-M1 is trained from scratch with a dedicated focus on enhancing medical capabilities. Our model is trained on 20 trillion tokens and incorporates a range of effective training methods that strike a balance between general capabilities and medical expertise. As a result, Baichuan-M1 not only performs strongly across general domains such as mathematics and coding but also excels in specialized medical fields. We have open-sourced Baichuan-M1-14B, a mini version of our model, which can be accessed through the following links.
Evaluating Entity Retrieval in Electronic Health Records: a Semantic Gap Perspective
Feb 10, 2025



Abstract:Entity retrieval plays a crucial role in the utilization of Electronic Health Records (EHRs) and is applied across a wide range of clinical practices. However, a comprehensive evaluation of this task is lacking due to the absence of a public benchmark. In this paper, we propose the development and release of a novel benchmark for evaluating entity retrieval in EHRs, with a particular focus on the semantic gap issue. Using discharge summaries from the MIMIC-III dataset, we incorporate ICD codes and prescription labels associated with the notes as queries, and annotate relevance judgments using GPT-4. In total, we use 1,000 patient notes, generate 1,246 queries, and provide over 77,000 relevance annotations. To offer the first assessment of the semantic gap, we introduce a novel classification system for relevance matches. Leveraging GPT-4, we categorize each relevant pair into one of five categories: string, synonym, abbreviation, hyponym, and implication. Using the proposed benchmark, we evaluate several retrieval methods, including BM25, query expansion, and state-of-the-art dense retrievers. Our findings show that BM25 provides a strong baseline but struggles with semantic matches. Query expansion significantly improves performance, though it slightly reduces string match capabilities. Dense retrievers outperform traditional methods, particularly for semantic matches, and general-domain dense retrievers often surpass those trained specifically in the biomedical domain.
GENIE: Generative Note Information Extraction model for structuring EHR data
Jan 30, 2025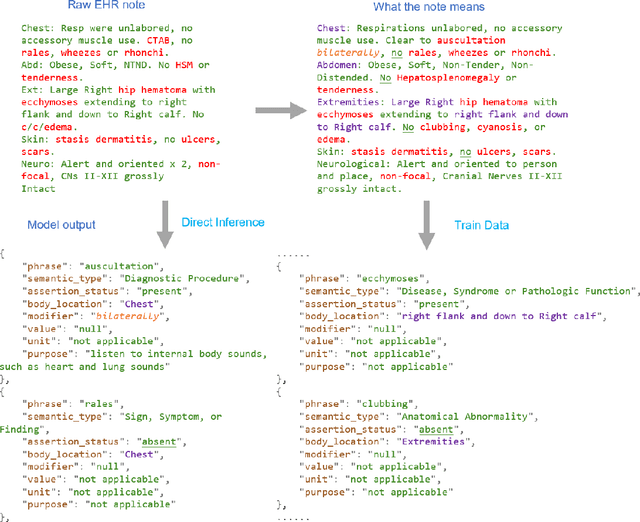
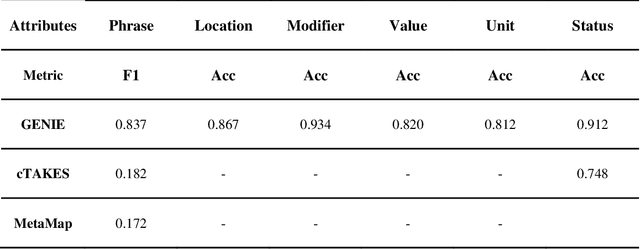
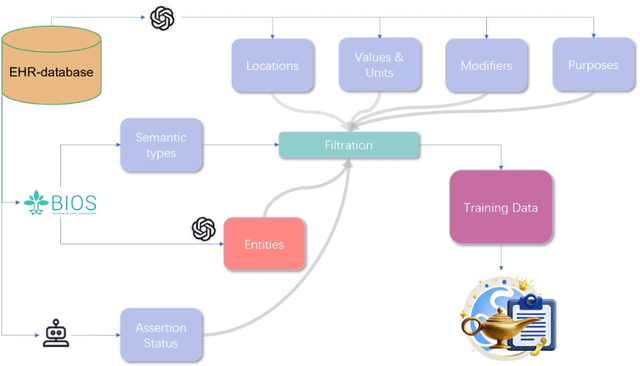
Abstract:Electronic Health Records (EHRs) hold immense potential for advancing healthcare, offering rich, longitudinal data that combines structured information with valuable insights from unstructured clinical notes. However, the unstructured nature of clinical text poses significant challenges for secondary applications. Traditional methods for structuring EHR free-text data, such as rule-based systems and multi-stage pipelines, are often limited by their time-consuming configurations and inability to adapt across clinical notes from diverse healthcare settings. Few systems provide a comprehensive attribute extraction for terminologies. While giant large language models (LLMs) like GPT-4 and LLaMA 405B excel at structuring tasks, they are slow, costly, and impractical for large-scale use. To overcome these limitations, we introduce GENIE, a Generative Note Information Extraction system that leverages LLMs to streamline the structuring of unstructured clinical text into usable data with standardized format. GENIE processes entire paragraphs in a single pass, extracting entities, assertion statuses, locations, modifiers, values, and purposes with high accuracy. Its unified, end-to-end approach simplifies workflows, reduces errors, and eliminates the need for extensive manual intervention. Using a robust data preparation pipeline and fine-tuned small scale LLMs, GENIE achieves competitive performance across multiple information extraction tasks, outperforming traditional tools like cTAKES and MetaMap and can handle extra attributes to be extracted. GENIE strongly enhances real-world applicability and scalability in healthcare systems. By open-sourcing the model and test data, we aim to encourage collaboration and drive further advancements in EHR structurization.
CoRTEx: Contrastive Learning for Representing Terms via Explanations with Applications on Constructing Biomedical Knowledge Graphs
Dec 13, 2023

Abstract:Objective: Biomedical Knowledge Graphs play a pivotal role in various biomedical research domains. Concurrently, term clustering emerges as a crucial step in constructing these knowledge graphs, aiming to identify synonymous terms. Due to a lack of knowledge, previous contrastive learning models trained with Unified Medical Language System (UMLS) synonyms struggle at clustering difficult terms and do not generalize well beyond UMLS terms. In this work, we leverage the world knowledge from Large Language Models (LLMs) and propose Contrastive Learning for Representing Terms via Explanations (CoRTEx) to enhance term representation and significantly improves term clustering. Materials and Methods: The model training involves generating explanations for a cleaned subset of UMLS terms using ChatGPT. We employ contrastive learning, considering term and explanation embeddings simultaneously, and progressively introduce hard negative samples. Additionally, a ChatGPT-assisted BIRCH algorithm is designed for efficient clustering of a new ontology. Results: We established a clustering test set and a hard negative test set, where our model consistently achieves the highest F1 score. With CoRTEx embeddings and the modified BIRCH algorithm, we grouped 35,580,932 terms from the Biomedical Informatics Ontology System (BIOS) into 22,104,559 clusters with O(N) queries to ChatGPT. Case studies highlight the model's efficacy in handling challenging samples, aided by information from explanations. Conclusion: By aligning terms to their explanations, CoRTEx demonstrates superior accuracy over benchmark models and robustness beyond its training set, and it is suitable for clustering terms for large-scale biomedical ontologies.
RAMM: Retrieval-augmented Biomedical Visual Question Answering with Multi-modal Pre-training
Mar 01, 2023
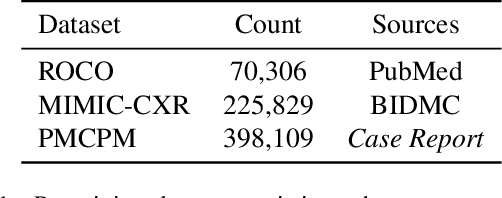
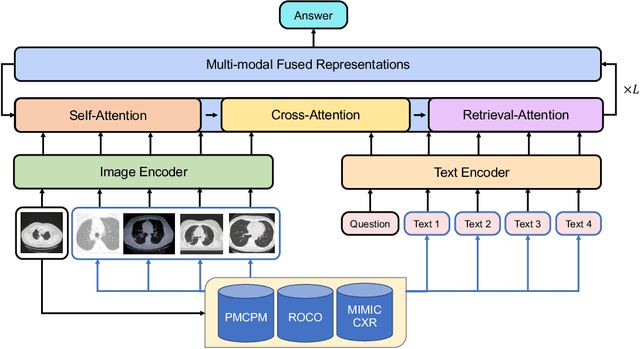
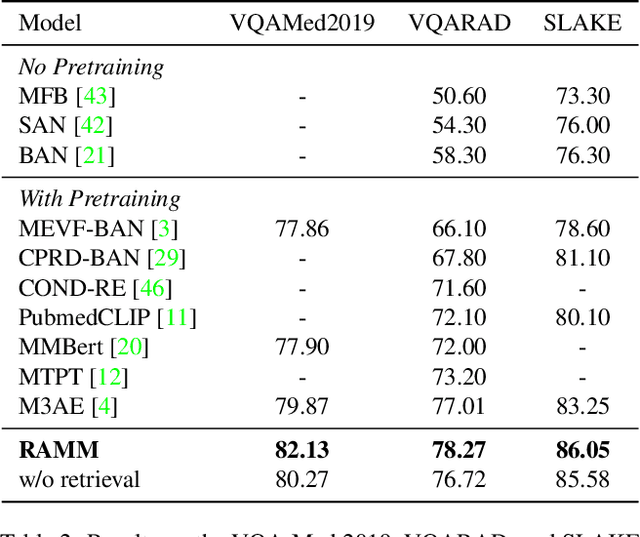
Abstract:Vision-and-language multi-modal pretraining and fine-tuning have shown great success in visual question answering (VQA). Compared to general domain VQA, the performance of biomedical VQA suffers from limited data. In this paper, we propose a retrieval-augmented pretrain-and-finetune paradigm named RAMM for biomedical VQA to overcome the data limitation issue. Specifically, we collect a new biomedical dataset named PMCPM which offers patient-based image-text pairs containing diverse patient situations from PubMed. Then, we pretrain the biomedical multi-modal model to learn visual and textual representation for image-text pairs and align these representations with image-text contrastive objective (ITC). Finally, we propose a retrieval-augmented method to better use the limited data. We propose to retrieve similar image-text pairs based on ITC from pretraining datasets and introduce a novel retrieval-attention module to fuse the representation of the image and the question with the retrieved images and texts. Experiments demonstrate that our retrieval-augmented pretrain-and-finetune paradigm obtains state-of-the-art performance on Med-VQA2019, Med-VQA2021, VQARAD, and SLAKE datasets. Further analysis shows that the proposed RAMM and PMCPM can enhance biomedical VQA performance compared with previous resources and methods. We will open-source our dataset, codes, and pretrained model.
BIOS: An Algorithmically Generated Biomedical Knowledge Graph
Mar 18, 2022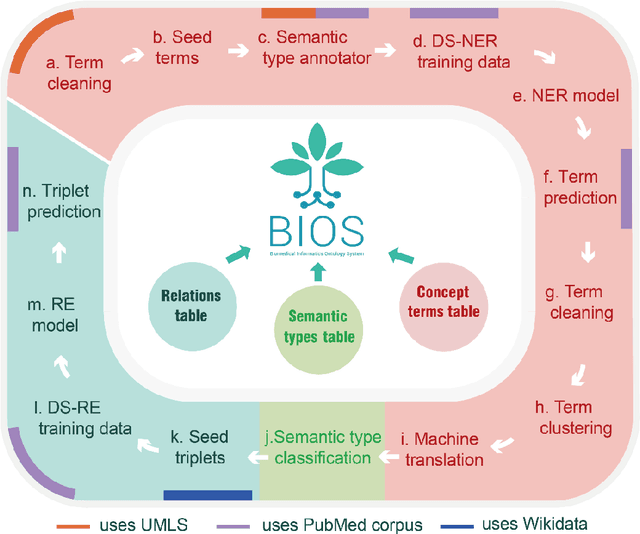
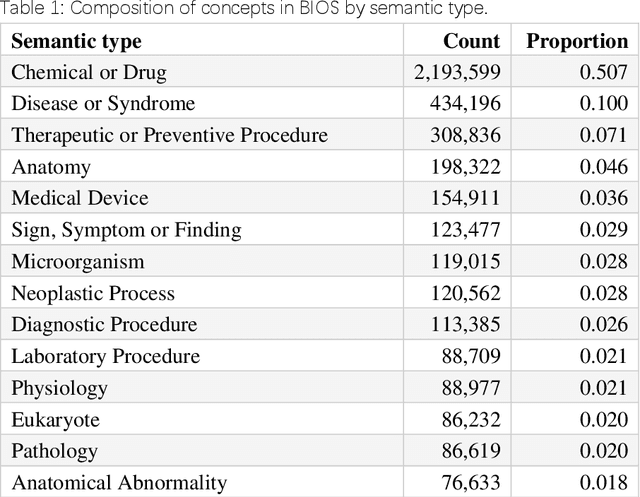

Abstract:Biomedical knowledge graphs (BioMedKGs) are essential infrastructures for biomedical and healthcare big data and artificial intelligence (AI), facilitating natural language processing, model development, and data exchange. For many decades, these knowledge graphs have been built via expert curation, which can no longer catch up with the speed of today's AI development, and a transition to algorithmically generated BioMedKGs is necessary. In this work, we introduce the Biomedical Informatics Ontology System (BIOS), the first large scale publicly available BioMedKG that is fully generated by machine learning algorithms. BIOS currently contains 4.1 million concepts, 7.4 million terms in two languages, and 7.3 million relation triplets. We introduce the methodology for developing BIOS, which covers curation of raw biomedical terms, computationally identifying synonymous terms and aggregating them to create concept nodes, semantic type classification of the concepts, relation identification, and biomedical machine translation. We provide statistics about the current content of BIOS and perform preliminary assessment for term quality, synonym grouping, and relation extraction. Results suggest that machine learning-based BioMedKG development is a totally viable solution for replacing traditional expert curation.
PMC-Patients: A Large-scale Dataset of Patient Notes and Relations Extracted from Case Reports in PubMed Central
Feb 28, 2022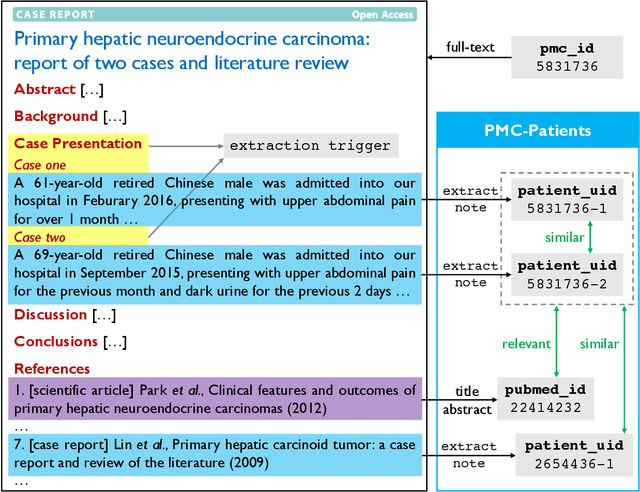
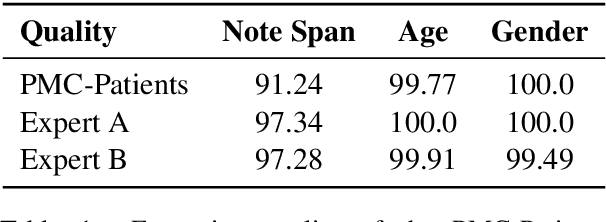
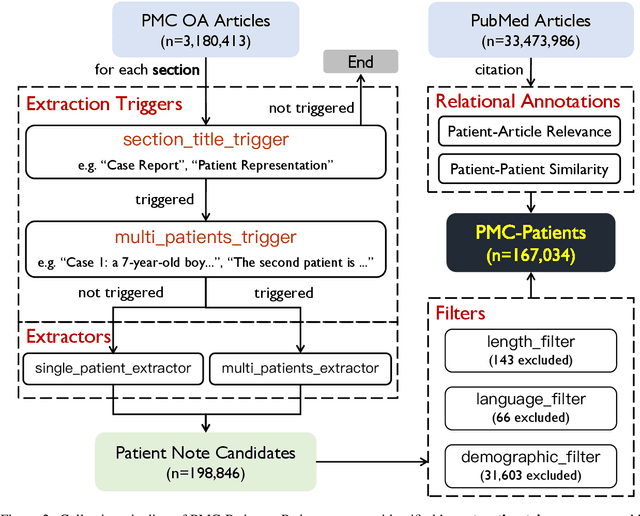
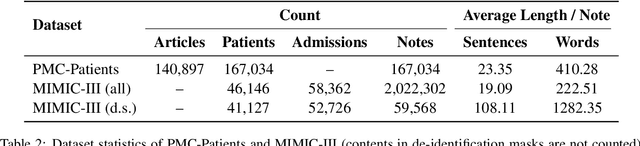
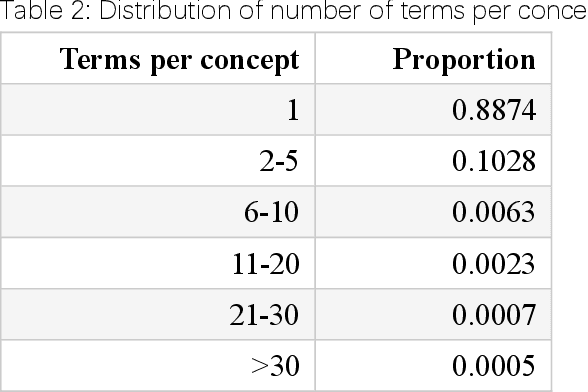
Abstract:We present PMC-Patients, a dataset consisting of 167k patient notes with 3.1M relevant article annotations and 293k similar patient annotations. The patient notes are extracted by identifying certain sections from case reports in PubMed Central, and those with at least CC BY-NC-SA license are re-distributed. Patient-article relevance and patient-patient similarity are defined by citation relationships in PubMed. We also perform four tasks with PMC-Patients to demonstrate its utility, including Patient Note Recognition (PNR), Patient-Patient Similarity (PPS), Patient-Patient Retrieval (PPR), and Patient-Article Retrieval (PAR). In summary, PMC-Patients provides the largest-scale patient notes with high quality, diverse conditions, easy access, and rich annotations.
CODER: Knowledge infused cross-lingual medical term embedding for term normalization
Nov 05, 2020



Abstract:We propose a novel medical term embedding method named CODER, which stands for mediCal knOwledge embeDded tErm Representation. CODER is designed for medical term normalization by providing close vector representations for terms that represent the same or similar concepts with multi-language support. CODER is trained on top of BERT (Devlin et al., 2018) with the innovation that token vector aggregation is trained using relations from the UMLS Metathesaurus (Bodenreider, 2004), which is a comprehensive medical knowledge graph with multi-language support. Training with relations injects medical knowledge into term embeddings and aims to provide better normalization performances and potentially better machine learning features. We evaluated CODER in term normalization, semantic similarity, and relation classification benchmarks, which showed that CODER outperformed various state-of-the-art biomedical word embeddings, concept embeddings, and contextual embeddings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge