Tianxi Cai
Learning Sequential Decisions from Multiple Sources via Group-Robust Markov Decision Processes
Feb 02, 2026Abstract:We often collect data from multiple sites (e.g., hospitals) that share common structure but also exhibit heterogeneity. This paper aims to learn robust sequential decision-making policies from such offline, multi-site datasets. To model cross-site uncertainty, we study distributionally robust MDPs with a group-linear structure: all sites share a common feature map, and both the transition kernels and expected reward functions are linear in these shared features. We introduce feature-wise (d-rectangular) uncertainty sets, which preserve tractable robust Bellman recursions while maintaining key cross-site structure. Building on this, we then develop an offline algorithm based on pessimistic value iteration that includes: (i) per-site ridge regression for Bellman targets, (ii) feature-wise worst-case (row-wise minimization) aggregation, and (iii) a data-dependent pessimism penalty computed from the diagonals of the inverse design matrices. We further propose a cluster-level extension that pools similar sites to improve sample efficiency, guided by prior knowledge of site similarity. Under a robust partial coverage assumption, we prove a suboptimality bound for the resulting policy. Overall, our framework addresses multi-site learning with heterogeneous data sources and provides a principled approach to robust planning without relying on strong state-action rectangularity assumptions.
Adversarial Drift-Aware Predictive Transfer: Toward Durable Clinical AI
Jan 21, 2026Abstract:Clinical AI systems frequently suffer performance decay post-deployment due to temporal data shifts, such as evolving populations, diagnostic coding updates (e.g., ICD-9 to ICD-10), and systemic shocks like the COVID-19 pandemic. Addressing this ``aging'' effect via frequent retraining is often impractical due to computational costs and privacy constraints. To overcome these hurdles, we introduce Adversarial Drift-Aware Predictive Transfer (ADAPT), a novel framework designed to confer durability against temporal drift with minimal retraining. ADAPT innovatively constructs an uncertainty set of plausible future models by combining historical source models and limited current data. By optimizing worst-case performance over this set, it balances current accuracy with robustness against degradation due to future drifts. Crucially, ADAPT requires only summary-level model estimators from historical periods, preserving data privacy and ensuring operational simplicity. Validated on longitudinal suicide risk prediction using electronic health records from Mass General Brigham (2005--2021) and Duke University Health Systems, ADAPT demonstrated superior stability across coding transitions and pandemic-induced shifts. By minimizing annual performance decay without labeling or retraining future data, ADAPT offers a scalable pathway for sustaining reliable AI in high-stakes healthcare environments.
Toward Global Large Language Models in Medicine
Jan 05, 2026Abstract:Despite continuous advances in medical technology, the global distribution of health care resources remains uneven. The development of large language models (LLMs) has transformed the landscape of medicine and holds promise for improving health care quality and expanding access to medical information globally. However, existing LLMs are primarily trained on high-resource languages, limiting their applicability in global medical scenarios. To address this gap, we constructed GlobMed, a large multilingual medical dataset, containing over 500,000 entries spanning 12 languages, including four low-resource languages. Building on this, we established GlobMed-Bench, which systematically assesses 56 state-of-the-art proprietary and open-weight LLMs across multiple multilingual medical tasks, revealing significant performance disparities across languages, particularly for low-resource languages. Additionally, we introduced GlobMed-LLMs, a suite of multilingual medical LLMs trained on GlobMed, with parameters ranging from 1.7B to 8B. GlobMed-LLMs achieved an average performance improvement of over 40% relative to baseline models, with a more than threefold increase in performance on low-resource languages. Together, these resources provide an important foundation for advancing the equitable development and application of LLMs globally, enabling broader language communities to benefit from technological advances.
PEHRT: A Common Pipeline for Harmonizing Electronic Health Record data for Translational Research
Sep 10, 2025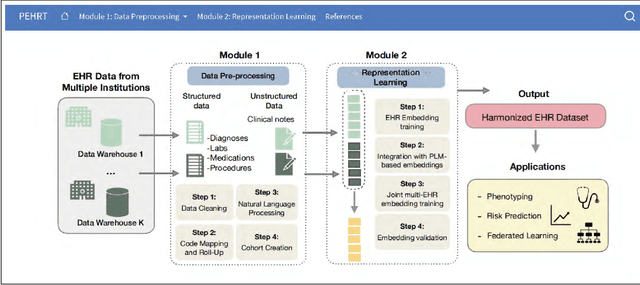
Abstract:Integrative analysis of multi-institutional Electronic Health Record (EHR) data enhances the reliability and generalizability of translational research by leveraging larger, more diverse patient cohorts and incorporating multiple data modalities. However, harmonizing EHR data across institutions poses major challenges due to data heterogeneity, semantic differences, and privacy concerns. To address these challenges, we introduce $\textit{PEHRT}$, a standardized pipeline for efficient EHR data harmonization consisting of two core modules: (1) data pre-processing and (2) representation learning. PEHRT maps EHR data to standard coding systems and uses advanced machine learning to generate research-ready datasets without requiring individual-level data sharing. Our pipeline is also data model agnostic and designed for streamlined execution across institutions based on our extensive real-world experience. We provide a complete suite of open source software, accompanied by a user-friendly tutorial, and demonstrate the utility of PEHRT in a variety of tasks using data from diverse healthcare systems.
One Patient, Many Contexts: Scaling Medical AI Through Contextual Intelligence
Jun 11, 2025

Abstract:Medical foundation models, including language models trained on clinical notes, vision-language models on medical images, and multimodal models on electronic health records, can summarize clinical notes, answer medical questions, and assist in decision-making. Adapting these models to new populations, specialties, or settings typically requires fine-tuning, careful prompting, or retrieval from knowledge bases. This can be impractical, and limits their ability to interpret unfamiliar inputs and adjust to clinical situations not represented during training. As a result, models are prone to contextual errors, where predictions appear reasonable but fail to account for critical patient-specific or contextual information. These errors stem from a fundamental limitation that current models struggle with: dynamically adjusting their behavior across evolving contexts of medical care. In this Perspective, we outline a vision for context-switching in medical AI: models that dynamically adapt their reasoning without retraining to new specialties, populations, workflows, and clinical roles. We envision context-switching AI to diagnose, manage, and treat a wide range of diseases across specialties and regions, and expand access to medical care.
Integrated Analysis for Electronic Health Records with Structured and Sporadic Missingness
Jun 10, 2025Abstract:Objectives: We propose a novel imputation method tailored for Electronic Health Records (EHRs) with structured and sporadic missingness. Such missingness frequently arises in the integration of heterogeneous EHR datasets for downstream clinical applications. By addressing these gaps, our method provides a practical solution for integrated analysis, enhancing data utility and advancing the understanding of population health. Materials and Methods: We begin by demonstrating structured and sporadic missing mechanisms in the integrated analysis of EHR data. Following this, we introduce a novel imputation framework, Macomss, specifically designed to handle structurally and heterogeneously occurring missing data. We establish theoretical guarantees for Macomss, ensuring its robustness in preserving the integrity and reliability of integrated analyses. To assess its empirical performance, we conduct extensive simulation studies that replicate the complex missingness patterns observed in real-world EHR systems, complemented by validation using EHR datasets from the Duke University Health System (DUHS). Results: Simulation studies show that our approach consistently outperforms existing imputation methods. Using datasets from three hospitals within DUHS, Macomss achieves the lowest imputation errors for missing data in most cases and provides superior or comparable downstream prediction performance compared to benchmark methods. Conclusions: We provide a theoretically guaranteed and practically meaningful method for imputing structured and sporadic missing data, enabling accurate and reliable integrated analysis across multiple EHR datasets. The proposed approach holds significant potential for advancing research in population health.
Semi-supervised Clustering Through Representation Learning of Large-scale EHR Data
May 27, 2025Abstract:Electronic Health Records (EHR) offer rich real-world data for personalized medicine, providing insights into disease progression, treatment responses, and patient outcomes. However, their sparsity, heterogeneity, and high dimensionality make them difficult to model, while the lack of standardized ground truth further complicates predictive modeling. To address these challenges, we propose SCORE, a semi-supervised representation learning framework that captures multi-domain disease profiles through patient embeddings. SCORE employs a Poisson-Adapted Latent factor Mixture (PALM) Model with pre-trained code embeddings to characterize codified features and extract meaningful patient phenotypes and embeddings. To handle the computational challenges of large-scale data, it introduces a hybrid Expectation-Maximization (EM) and Gaussian Variational Approximation (GVA) algorithm, leveraging limited labeled data to refine estimates on a vast pool of unlabeled samples. We theoretically establish the convergence of this hybrid approach, quantify GVA errors, and derive SCORE's error rate under diverging embedding dimensions. Our analysis shows that incorporating unlabeled data enhances accuracy and reduces sensitivity to label scarcity. Extensive simulations confirm SCORE's superior finite-sample performance over existing methods. Finally, we apply SCORE to predict disability status for patients with multiple sclerosis (MS) using partially labeled EHR data, demonstrating that it produces more informative and predictive patient embeddings for multiple MS-related conditions compared to existing approaches.
A Theoretical Framework for Prompt Engineering: Approximating Smooth Functions with Transformer Prompts
Mar 26, 2025Abstract:Prompt engineering has emerged as a powerful technique for guiding large language models (LLMs) toward desired responses, significantly enhancing their performance across diverse tasks. Beyond their role as static predictors, LLMs increasingly function as intelligent agents, capable of reasoning, decision-making, and adapting dynamically to complex environments. However, the theoretical underpinnings of prompt engineering remain largely unexplored. In this paper, we introduce a formal framework demonstrating that transformer models, when provided with carefully designed prompts, can act as a configurable computational system by emulating a ``virtual'' neural network during inference. Specifically, input prompts effectively translate into the corresponding network configuration, enabling LLMs to adjust their internal computations dynamically. Building on this construction, we establish an approximation theory for $\beta$-times differentiable functions, proving that transformers can approximate such functions with arbitrary precision when guided by appropriately structured prompts. Moreover, our framework provides theoretical justification for several empirically successful prompt engineering techniques, including the use of longer, structured prompts, filtering irrelevant information, enhancing prompt token diversity, and leveraging multi-agent interactions. By framing LLMs as adaptable agents rather than static models, our findings underscore their potential for autonomous reasoning and problem-solving, paving the way for more robust and theoretically grounded advancements in prompt engineering and AI agent design.
Representation Learning to Advance Multi-institutional Studies with Electronic Health Record Data
Feb 12, 2025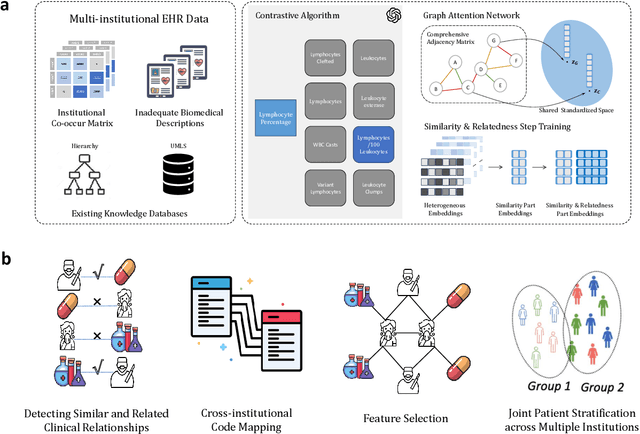
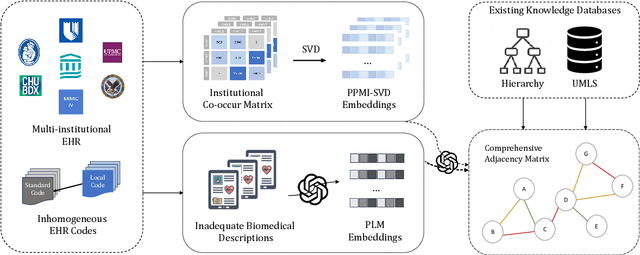
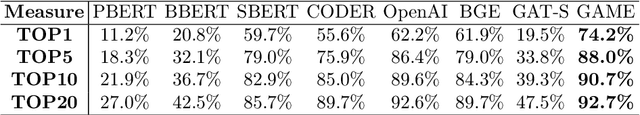
Abstract:The adoption of EHRs has expanded opportunities to leverage data-driven algorithms in clinical care and research. A major bottleneck in effectively conducting multi-institutional EHR studies is the data heterogeneity across systems with numerous codes that either do not exist or represent different clinical concepts across institutions. The need for data privacy further limits the feasibility of including multi-institutional patient-level data required to study similarities and differences across patient subgroups. To address these challenges, we developed the GAME algorithm. Tested and validated across 7 institutions and 2 languages, GAME integrates data in several levels: (1) at the institutional level with knowledge graphs to establish relationships between codes and existing knowledge sources, providing the medical context for standard codes and their relationship to each other; (2) between institutions, leveraging language models to determine the relationships between institution-specific codes with established standard codes; and (3) quantifying the strength of the relationships between codes using a graph attention network. Jointly trained embeddings are created using transfer and federated learning to preserve data privacy. In this study, we demonstrate the applicability of GAME in selecting relevant features as inputs for AI-driven algorithms in a range of conditions, e.g., heart failure, rheumatoid arthritis. We then highlight the application of GAME harmonized multi-institutional EHR data in a study of Alzheimer's disease outcomes and suicide risk among patients with mental health disorders, without sharing patient-level data outside individual institutions.
GENIE: Generative Note Information Extraction model for structuring EHR data
Jan 30, 2025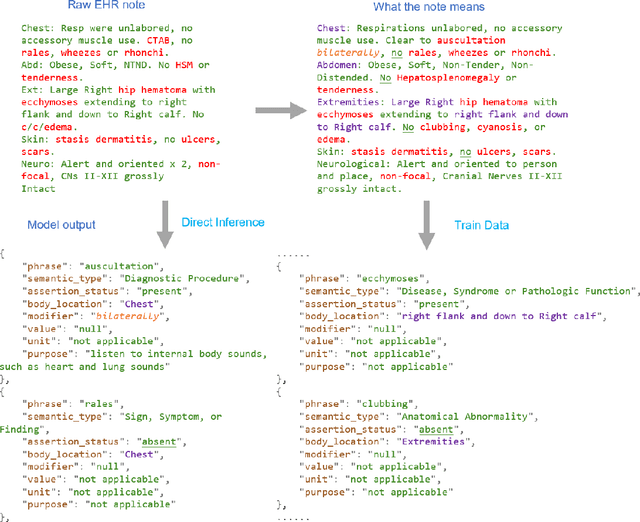
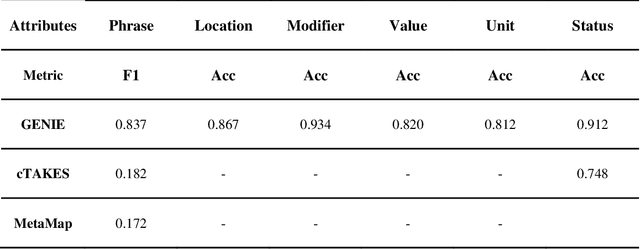
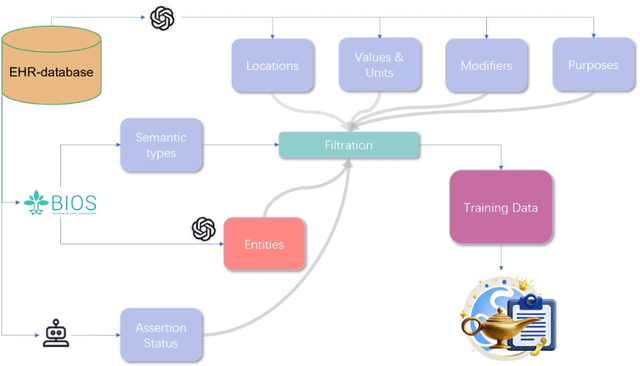
Abstract:Electronic Health Records (EHRs) hold immense potential for advancing healthcare, offering rich, longitudinal data that combines structured information with valuable insights from unstructured clinical notes. However, the unstructured nature of clinical text poses significant challenges for secondary applications. Traditional methods for structuring EHR free-text data, such as rule-based systems and multi-stage pipelines, are often limited by their time-consuming configurations and inability to adapt across clinical notes from diverse healthcare settings. Few systems provide a comprehensive attribute extraction for terminologies. While giant large language models (LLMs) like GPT-4 and LLaMA 405B excel at structuring tasks, they are slow, costly, and impractical for large-scale use. To overcome these limitations, we introduce GENIE, a Generative Note Information Extraction system that leverages LLMs to streamline the structuring of unstructured clinical text into usable data with standardized format. GENIE processes entire paragraphs in a single pass, extracting entities, assertion statuses, locations, modifiers, values, and purposes with high accuracy. Its unified, end-to-end approach simplifies workflows, reduces errors, and eliminates the need for extensive manual intervention. Using a robust data preparation pipeline and fine-tuned small scale LLMs, GENIE achieves competitive performance across multiple information extraction tasks, outperforming traditional tools like cTAKES and MetaMap and can handle extra attributes to be extracted. GENIE strongly enhances real-world applicability and scalability in healthcare systems. By open-sourcing the model and test data, we aim to encourage collaboration and drive further advancements in EHR structurization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge