Doudou Zhou
Department of Statistics and Data Science, National University of Singapore, Singapore
Learning Sequential Decisions from Multiple Sources via Group-Robust Markov Decision Processes
Feb 02, 2026Abstract:We often collect data from multiple sites (e.g., hospitals) that share common structure but also exhibit heterogeneity. This paper aims to learn robust sequential decision-making policies from such offline, multi-site datasets. To model cross-site uncertainty, we study distributionally robust MDPs with a group-linear structure: all sites share a common feature map, and both the transition kernels and expected reward functions are linear in these shared features. We introduce feature-wise (d-rectangular) uncertainty sets, which preserve tractable robust Bellman recursions while maintaining key cross-site structure. Building on this, we then develop an offline algorithm based on pessimistic value iteration that includes: (i) per-site ridge regression for Bellman targets, (ii) feature-wise worst-case (row-wise minimization) aggregation, and (iii) a data-dependent pessimism penalty computed from the diagonals of the inverse design matrices. We further propose a cluster-level extension that pools similar sites to improve sample efficiency, guided by prior knowledge of site similarity. Under a robust partial coverage assumption, we prove a suboptimality bound for the resulting policy. Overall, our framework addresses multi-site learning with heterogeneous data sources and provides a principled approach to robust planning without relying on strong state-action rectangularity assumptions.
A Judge-Aware Ranking Framework for Evaluating Large Language Models without Ground Truth
Jan 29, 2026Abstract:Evaluating large language models (LLMs) on open-ended tasks without ground-truth labels is increasingly done via the LLM-as-a-judge paradigm. A critical but under-modeled issue is that judge LLMs differ substantially in reliability; treating all judges equally can yield biased leaderboards and misleading uncertainty estimates. More data can make evaluation more confidently wrong under misspecified aggregation. We propose a judge-aware ranking framework that extends the Bradley-Terry-Luce model by introducing judge-specific discrimination parameters, jointly estimating latent model quality and judge reliability from pairwise comparisons without reference labels. We establish identifiability up to natural normalizations and prove consistency and asymptotic normality of the maximum likelihood estimator, enabling confidence intervals for score differences and rank comparisons. Across multiple public benchmarks and a newly collected dataset, our method improves agreement with human preferences, achieves higher data efficiency than unweighted baselines, and produces calibrated uncertainty quantification for LLM rankings.
From Hawkes Processes to Attention: Time-Modulated Mechanisms for Event Sequences
Jan 14, 2026Abstract:Marked Temporal Point Processes (MTPPs) arise naturally in medical, social, commercial, and financial domains. However, existing Transformer-based methods mostly inject temporal information only via positional encodings, relying on shared or parametric decay structures, which limits their ability to capture heterogeneous and type-specific temporal effects. Inspired by this observation, we derive a novel attention operator called Hawkes Attention from the multivariate Hawkes process theory for MTPP, using learnable per-type neural kernels to modulate query, key and value projections, thereby replacing the corresponding parts in the traditional attention. Benefited from the design, Hawkes Attention unifies event timing and content interaction, learning both the time-relevant behavior and type-specific excitation patterns from the data. The experimental results show that our method achieves better performance compared to the baselines. In addition to the general MTPP, our attention mechanism can also be easily applied to specific temporal structures, such as time series forecasting.
RELEAP: Reinforcement-Enhanced Label-Efficient Active Phenotyping for Electronic Health Records
Nov 08, 2025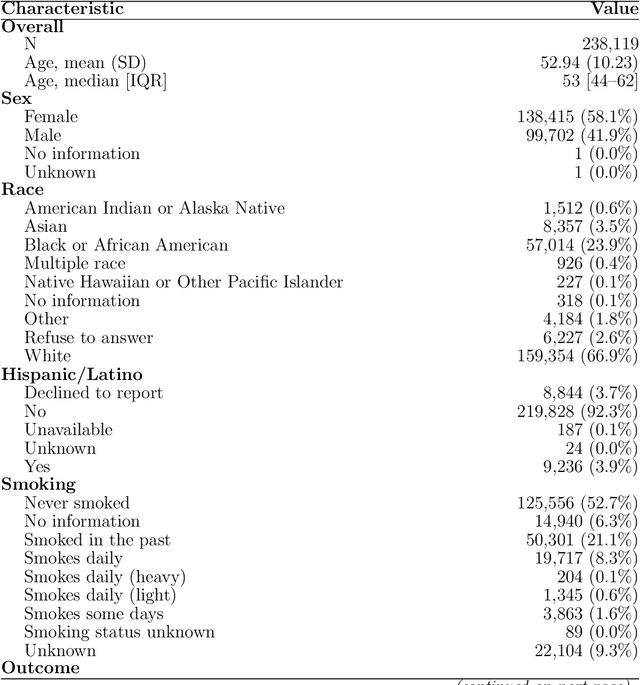
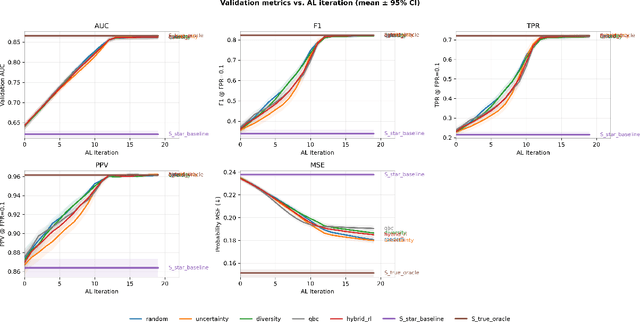

Abstract:Objective: Electronic health record (EHR) phenotyping often relies on noisy proxy labels, which undermine the reliability of downstream risk prediction. Active learning can reduce annotation costs, but most rely on fixed heuristics and do not ensure that phenotype refinement improves prediction performance. Our goal was to develop a framework that directly uses downstream prediction performance as feedback to guide phenotype correction and sample selection under constrained labeling budgets. Materials and Methods: We propose Reinforcement-Enhanced Label-Efficient Active Phenotyping (RELEAP), a reinforcement learning-based active learning framework. RELEAP adaptively integrates multiple querying strategies and, unlike prior methods, updates its policy based on feedback from downstream models. We evaluated RELEAP on a de-identified Duke University Health System (DUHS) cohort (2014-2024) for incident lung cancer risk prediction, using logistic regression and penalized Cox survival models. Performance was benchmarked against noisy-label baselines and single-strategy active learning. Results: RELEAP consistently outperformed all baselines. Logistic AUC increased from 0.774 to 0.805 and survival C-index from 0.718 to 0.752. Using downstream performance as feedback, RELEAP produced smoother and more stable gains than heuristic methods under the same labeling budget. Discussion: By linking phenotype refinement to prediction outcomes, RELEAP learns which samples most improve downstream discrimination and calibration, offering a more principled alternative to fixed active learning rules. Conclusion: RELEAP optimizes phenotype correction through downstream feedback, offering a scalable, label-efficient paradigm that reduces manual chart review and enhances the reliability of EHR-based risk prediction.
PEHRT: A Common Pipeline for Harmonizing Electronic Health Record data for Translational Research
Sep 10, 2025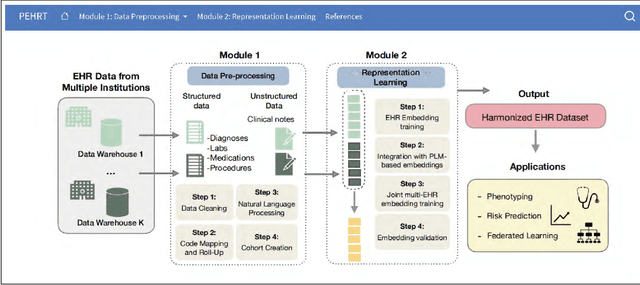
Abstract:Integrative analysis of multi-institutional Electronic Health Record (EHR) data enhances the reliability and generalizability of translational research by leveraging larger, more diverse patient cohorts and incorporating multiple data modalities. However, harmonizing EHR data across institutions poses major challenges due to data heterogeneity, semantic differences, and privacy concerns. To address these challenges, we introduce $\textit{PEHRT}$, a standardized pipeline for efficient EHR data harmonization consisting of two core modules: (1) data pre-processing and (2) representation learning. PEHRT maps EHR data to standard coding systems and uses advanced machine learning to generate research-ready datasets without requiring individual-level data sharing. Our pipeline is also data model agnostic and designed for streamlined execution across institutions based on our extensive real-world experience. We provide a complete suite of open source software, accompanied by a user-friendly tutorial, and demonstrate the utility of PEHRT in a variety of tasks using data from diverse healthcare systems.
Single Index Bandits: Generalized Linear Contextual Bandits with Unknown Reward Functions
Jun 15, 2025Abstract:Generalized linear bandits have been extensively studied due to their broad applicability in real-world online decision-making problems. However, these methods typically assume that the expected reward function is known to the users, an assumption that is often unrealistic in practice. Misspecification of this link function can lead to the failure of all existing algorithms. In this work, we address this critical limitation by introducing a new problem of generalized linear bandits with unknown reward functions, also known as single index bandits. We first consider the case where the unknown reward function is monotonically increasing, and propose two novel and efficient algorithms, STOR and ESTOR, that achieve decent regrets under standard assumptions. Notably, our ESTOR can obtain the nearly optimal regret bound $\tilde{O}_T(\sqrt{T})$ in terms of the time horizon $T$. We then extend our methods to the high-dimensional sparse setting and show that the same regret rate can be attained with the sparsity index. Next, we introduce GSTOR, an algorithm that is agnostic to general reward functions, and establish regret bounds under a Gaussian design assumption. Finally, we validate the efficiency and effectiveness of our algorithms through experiments on both synthetic and real-world datasets.
Wasserstein Transfer Learning
May 23, 2025Abstract:Transfer learning is a powerful paradigm for leveraging knowledge from source domains to enhance learning in a target domain. However, traditional transfer learning approaches often focus on scalar or multivariate data within Euclidean spaces, limiting their applicability to complex data structures such as probability distributions. To address this, we introduce a novel framework for transfer learning in regression models, where outputs are probability distributions residing in the Wasserstein space. When the informative subset of transferable source domains is known, we propose an estimator with provable asymptotic convergence rates, quantifying the impact of domain similarity on transfer efficiency. For cases where the informative subset is unknown, we develop a data-driven transfer learning procedure designed to mitigate negative transfer. The proposed methods are supported by rigorous theoretical analysis and are validated through extensive simulations and real-world applications.
Toward Fair Federated Learning under Demographic Disparities and Data Imbalance
May 14, 2025Abstract:Ensuring fairness is critical when applying artificial intelligence to high-stakes domains such as healthcare, where predictive models trained on imbalanced and demographically skewed data risk exacerbating existing disparities. Federated learning (FL) enables privacy-preserving collaboration across institutions, but remains vulnerable to both algorithmic bias and subgroup imbalance - particularly when multiple sensitive attributes intersect. We propose FedIDA (Fed erated Learning for Imbalance and D isparity A wareness), a framework-agnostic method that combines fairness-aware regularization with group-conditional oversampling. FedIDA supports multiple sensitive attributes and heterogeneous data distributions without altering the convergence behavior of the underlying FL algorithm. We provide theoretical analysis establishing fairness improvement bounds using Lipschitz continuity and concentration inequalities, and show that FedIDA reduces the variance of fairness metrics across test sets. Empirical results on both benchmark and real-world clinical datasets confirm that FedIDA consistently improves fairness while maintaining competitive predictive performance, demonstrating its effectiveness for equitable and privacy-preserving modeling in healthcare. The source code is available on GitHub.
SIM-Shapley: A Stable and Computationally Efficient Approach to Shapley Value Approximation
May 13, 2025



Abstract:Explainable artificial intelligence (XAI) is essential for trustworthy machine learning (ML), particularly in high-stakes domains such as healthcare and finance. Shapley value (SV) methods provide a principled framework for feature attribution in complex models but incur high computational costs, limiting their scalability in high-dimensional settings. We propose Stochastic Iterative Momentum for Shapley Value Approximation (SIM-Shapley), a stable and efficient SV approximation method inspired by stochastic optimization. We analyze variance theoretically, prove linear $Q$-convergence, and demonstrate improved empirical stability and low bias in practice on real-world datasets. In our numerical experiments, SIM-Shapley reduces computation time by up to 85% relative to state-of-the-art baselines while maintaining comparable feature attribution quality. Beyond feature attribution, our stochastic mini-batch iterative framework extends naturally to a broader class of sample average approximation problems, offering a new avenue for improving computational efficiency with stability guarantees. Code is publicly available at https://github.com/nliulab/SIM-Shapley.
Representation Learning to Advance Multi-institutional Studies with Electronic Health Record Data
Feb 12, 2025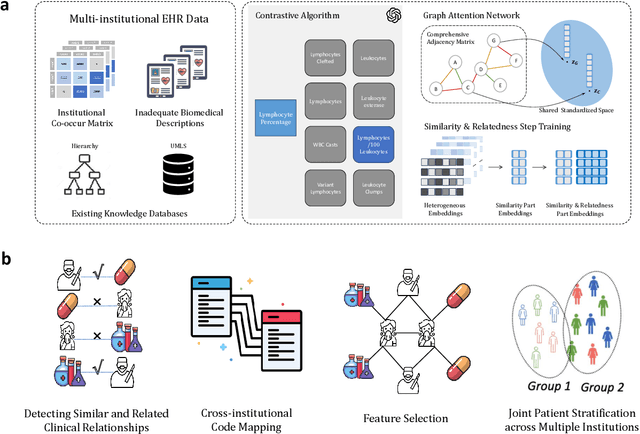
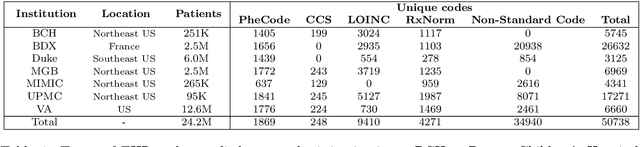
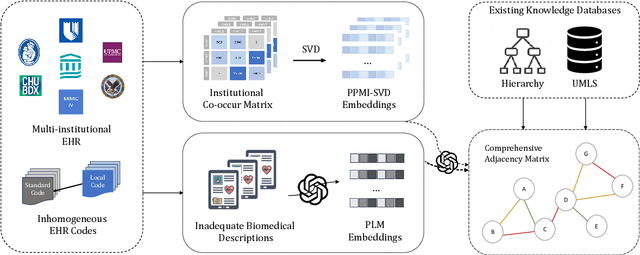
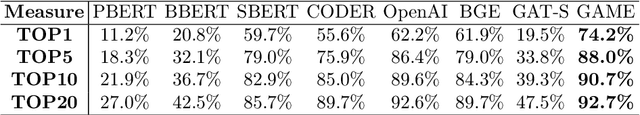
Abstract:The adoption of EHRs has expanded opportunities to leverage data-driven algorithms in clinical care and research. A major bottleneck in effectively conducting multi-institutional EHR studies is the data heterogeneity across systems with numerous codes that either do not exist or represent different clinical concepts across institutions. The need for data privacy further limits the feasibility of including multi-institutional patient-level data required to study similarities and differences across patient subgroups. To address these challenges, we developed the GAME algorithm. Tested and validated across 7 institutions and 2 languages, GAME integrates data in several levels: (1) at the institutional level with knowledge graphs to establish relationships between codes and existing knowledge sources, providing the medical context for standard codes and their relationship to each other; (2) between institutions, leveraging language models to determine the relationships between institution-specific codes with established standard codes; and (3) quantifying the strength of the relationships between codes using a graph attention network. Jointly trained embeddings are created using transfer and federated learning to preserve data privacy. In this study, we demonstrate the applicability of GAME in selecting relevant features as inputs for AI-driven algorithms in a range of conditions, e.g., heart failure, rheumatoid arthritis. We then highlight the application of GAME harmonized multi-institutional EHR data in a study of Alzheimer's disease outcomes and suicide risk among patients with mental health disorders, without sharing patient-level data outside individual institutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge