Yucong Lin
Scale-Aware Relay and Scale-Adaptive Loss for Tiny Object Detection in Aerial Images
Nov 13, 2025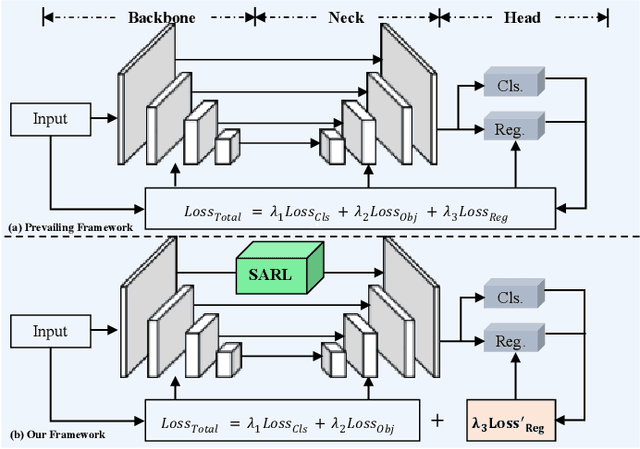
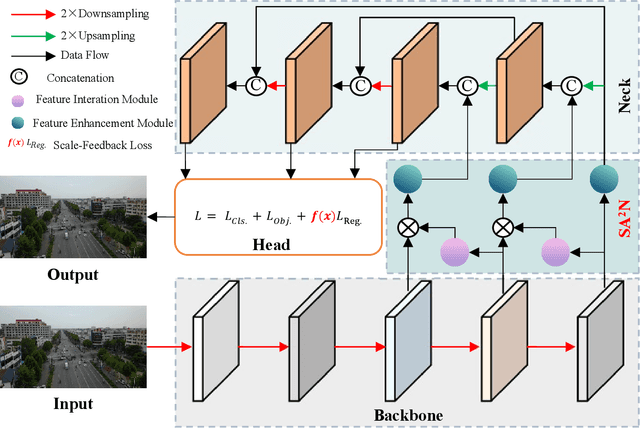
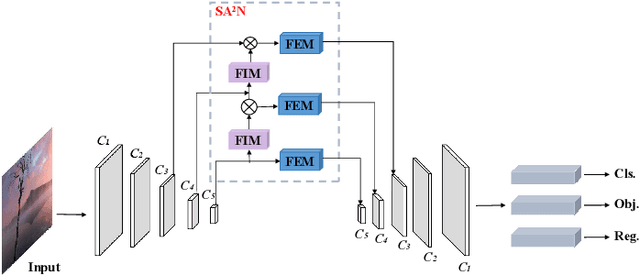
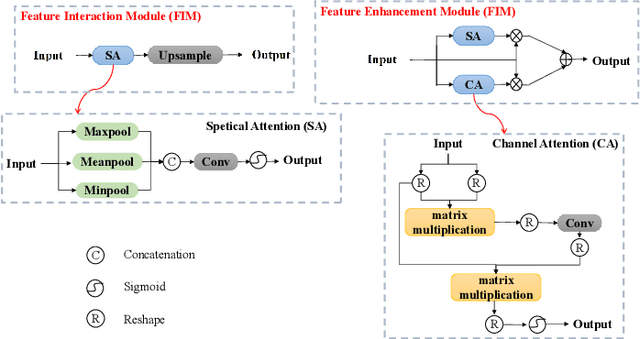
Abstract:Recently, despite the remarkable advancements in object detection, modern detectors still struggle to detect tiny objects in aerial images. One key reason is that tiny objects carry limited features that are inevitably degraded or lost during long-distance network propagation. Another is that smaller objects receive disproportionately greater regression penalties than larger ones during training. To tackle these issues, we propose a Scale-Aware Relay Layer (SARL) and a Scale-Adaptive Loss (SAL) for tiny object detection, both of which are seamlessly compatible with the top-performing frameworks. Specifically, SARL employs a cross-scale spatial-channel attention to progressively enrich the meaningful features of each layer and strengthen the cross-layer feature sharing. SAL reshapes the vanilla IoU-based losses so as to dynamically assign lower weights to larger objects. This loss is able to focus training on tiny objects while reducing the influence on large objects. Extensive experiments are conducted on three benchmarks (\textit{i.e.,} AI-TOD, DOTA-v2.0 and VisDrone2019), and the results demonstrate that the proposed method boosts the generalization ability by 5.5\% Average Precision (AP) when embedded in YOLOv5 (anchor-based) and YOLOx (anchor-free) baselines. Moreover, it also promotes the robust performance with 29.0\% AP on the real-world noisy dataset (\textit{i.e.,} AI-TOD-v2.0).
Sculpting Margin Penalty: Intra-Task Adapter Merging and Classifier Calibration for Few-Shot Class-Incremental Learning
Aug 07, 2025Abstract:Real-world applications often face data privacy constraints and high acquisition costs, making the assumption of sufficient training data in incremental tasks unrealistic and leading to significant performance degradation in class-incremental learning. Forward-compatible learning, which prospectively prepares for future tasks during base task training, has emerged as a promising solution for Few-Shot Class-Incremental Learning (FSCIL). However, existing methods still struggle to balance base-class discriminability and new-class generalization. Moreover, limited access to original data during incremental tasks often results in ambiguous inter-class decision boundaries. To address these challenges, we propose SMP (Sculpting Margin Penalty), a novel FSCIL method that strategically integrates margin penalties at different stages within the parameter-efficient fine-tuning paradigm. Specifically, we introduce the Margin-aware Intra-task Adapter Merging (MIAM) mechanism for base task learning. MIAM trains two sets of low-rank adapters with distinct classification losses: one with a margin penalty to enhance base-class discriminability, and the other without margin constraints to promote generalization to future new classes. These adapters are then adaptively merged to improve forward compatibility. For incremental tasks, we propose a Margin Penalty-based Classifier Calibration (MPCC) strategy to refine decision boundaries by fine-tuning classifiers on all seen classes' embeddings with a margin penalty. Extensive experiments on CIFAR100, ImageNet-R, and CUB200 demonstrate that SMP achieves state-of-the-art performance in FSCIL while maintaining a better balance between base and new classes.
MetaFE-DE: Learning Meta Feature Embedding for Depth Estimation from Monocular Endoscopic Images
Feb 05, 2025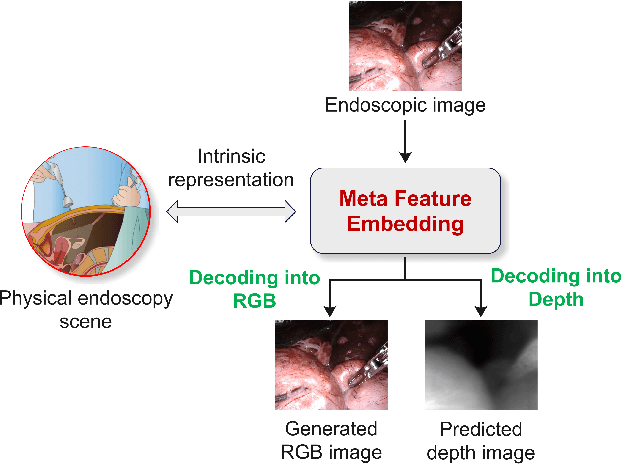
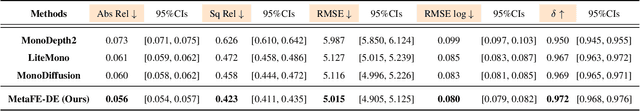
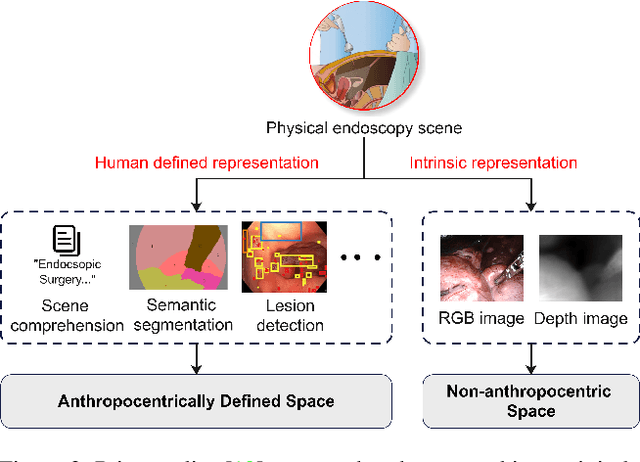
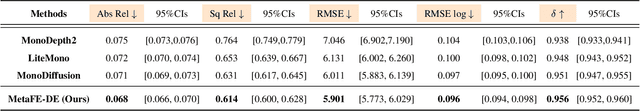
Abstract:Depth estimation from monocular endoscopic images presents significant challenges due to the complexity of endoscopic surgery, such as irregular shapes of human soft tissues, as well as variations in lighting conditions. Existing methods primarily estimate the depth information from RGB images directly, and often surffer the limited interpretability and accuracy. Given that RGB and depth images are two views of the same endoscopic surgery scene, in this paper, we introduce a novel concept referred as ``meta feature embedding (MetaFE)", in which the physical entities (e.g., tissues and surgical instruments) of endoscopic surgery are represented using the shared features that can be alternatively decoded into RGB or depth image. With this concept, we propose a two-stage self-supervised learning paradigm for the monocular endoscopic depth estimation. In the first stage, we propose a temporal representation learner using diffusion models, which are aligned with the spatial information through the cross normalization to construct the MetaFE. In the second stage, self-supervised monocular depth estimation with the brightness calibration is applied to decode the meta features into the depth image. Extensive evaluation on diverse endoscopic datasets demonstrates that our approach outperforms the state-of-the-art method in depth estimation, achieving superior accuracy and generalization. The source code will be publicly available.
Efficient Non-Exemplar Class-Incremental Learning with Retrospective Feature Synthesis
Nov 03, 2024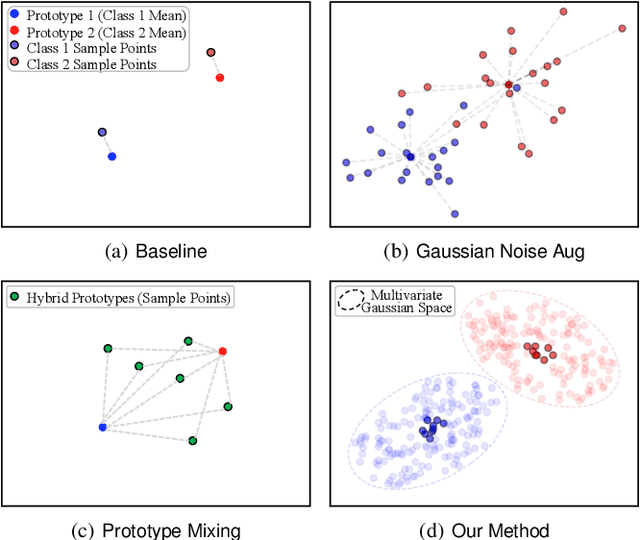
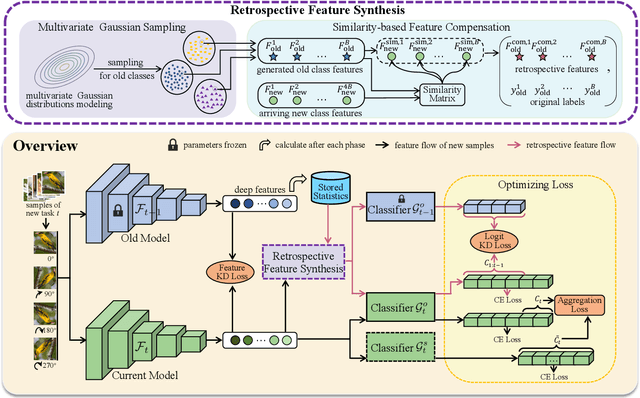
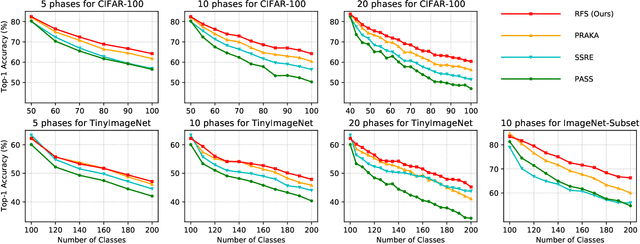
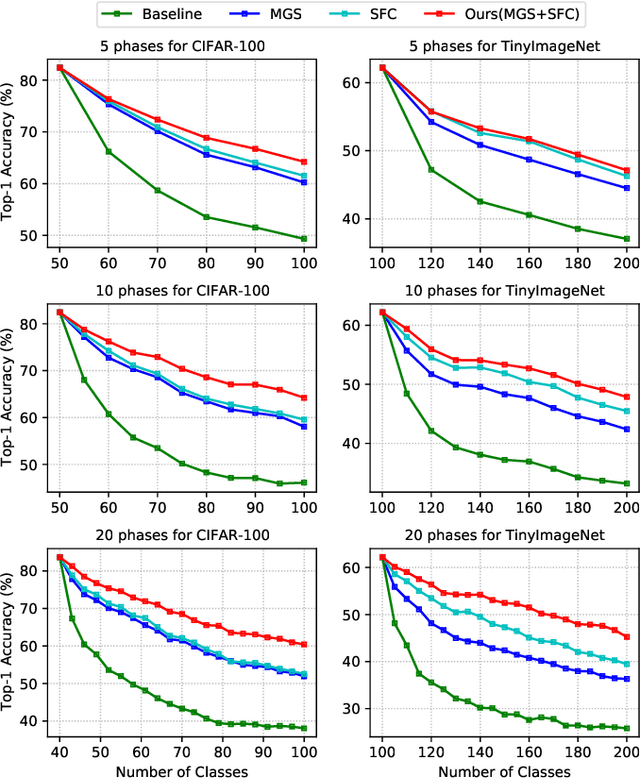
Abstract:Despite the outstanding performance in many individual tasks, deep neural networks suffer from catastrophic forgetting when learning from continuous data streams in real-world scenarios. Current Non-Exemplar Class-Incremental Learning (NECIL) methods mitigate forgetting by storing a single prototype per class, which serves to inject previous information when sequentially learning new classes. However, these stored prototypes or their augmented variants often fail to simultaneously capture spatial distribution diversity and precision needed for representing old classes. Moreover, as the model acquires new knowledge, these prototypes gradually become outdated, making them less effective. To overcome these limitations, we propose a more efficient NECIL method that replaces prototypes with synthesized retrospective features for old classes. Specifically, we model each old class's feature space using a multivariate Gaussian distribution and generate deep representations by sampling from high-likelihood regions. Additionally, we introduce a similarity-based feature compensation mechanism that integrates generated old class features with similar new class features to synthesize robust retrospective representations. These retrospective features are then incorporated into our incremental learning framework to preserve the decision boundaries of previous classes while learning new ones. Extensive experiments on CIFAR-100, TinyImageNet, and ImageNet-Subset demonstrate that our method significantly improves the efficiency of non-exemplar class-incremental learning and achieves state-of-the-art performance.
Hierarchical Pretraining for Biomedical Term Embeddings
Jul 01, 2023


Abstract:Electronic health records (EHR) contain narrative notes that provide extensive details on the medical condition and management of patients. Natural language processing (NLP) of clinical notes can use observed frequencies of clinical terms as predictive features for downstream applications such as clinical decision making and patient trajectory prediction. However, due to the vast number of highly similar and related clinical concepts, a more effective modeling strategy is to represent clinical terms as semantic embeddings via representation learning and use the low dimensional embeddings as feature vectors for predictive modeling. To achieve efficient representation, fine-tuning pretrained language models with biomedical knowledge graphs may generate better embeddings for biomedical terms than those from standard language models alone. These embeddings can effectively discriminate synonymous pairs of from those that are unrelated. However, they often fail to capture different degrees of similarity or relatedness for concepts that are hierarchical in nature. To overcome this limitation, we propose HiPrBERT, a novel biomedical term representation model trained on additionally complied data that contains hierarchical structures for various biomedical terms. We modify an existing contrastive loss function to extract information from these hierarchies. Our numerical experiments demonstrate that HiPrBERT effectively learns the pair-wise distance from hierarchical information, resulting in a substantially more informative embeddings for further biomedical applications
High-Resolution Boundary Detection for Medical Image Segmentation with Piece-Wise Two-Sample T-Test Augmented Loss
Nov 04, 2022



Abstract:Deep learning methods have contributed substantially to the rapid advancement of medical image segmentation, the quality of which relies on the suitable design of loss functions. Popular loss functions, including the cross-entropy and dice losses, often fall short of boundary detection, thereby limiting high-resolution downstream applications such as automated diagnoses and procedures. We developed a novel loss function that is tailored to reflect the boundary information to enhance the boundary detection. As the contrast between segmentation and background regions along the classification boundary naturally induces heterogeneity over the pixels, we propose the piece-wise two-sample t-test augmented (PTA) loss that is infused with the statistical test for such heterogeneity. We demonstrate the improved boundary detection power of the PTA loss compared to benchmark losses without a t-test component.
Knowledge Graph Enhanced Relation Extraction Datasets
Oct 19, 2022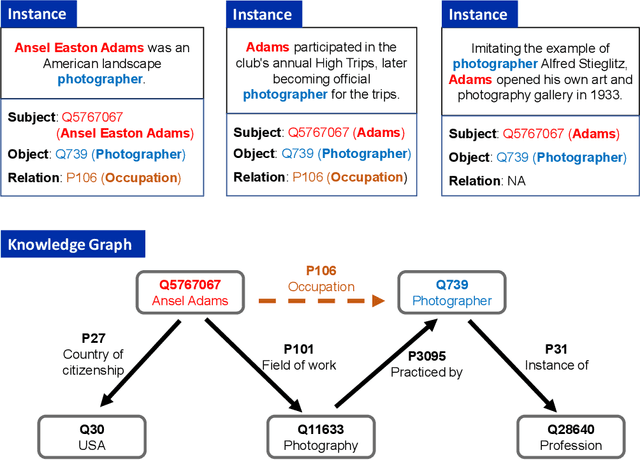


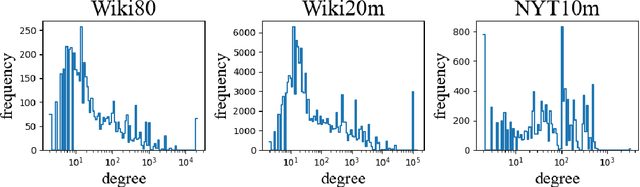
Abstract:Knowledge-enhanced methods that take advantage of auxiliary knowledge graphs recently emerged in relation extraction, and they surpass traditional text-based relation extraction methods. However, there are no unified public benchmarks that currently involve evidence sentences and knowledge graphs for knowledge-enhanced relation extraction. To combat these issues, we propose KGRED, a knowledge graph enhanced relation extraction dataset with features as follows: (1) the benchmarks are based on widely-used distantly supervised relation extraction datasets; (2) we refine these existing datasets to improve the data quality, and we also construct auxiliary knowledge graphs for these existing datasets through entity linking to support knowledge-enhanced relation extraction tasks; (3) with the new benchmarks we curated, we build baselines in two popular relation extraction settings including sentence-level and bag-level relation extraction, and we also make comparisons among the latest knowledge-enhanced relation extraction methods. KGRED provides high-quality relation extraction datasets with auxiliary knowledge graphs for evaluating the performance of knowledge-enhanced relation extraction methods. Meanwhile, our experiments on KGRED reveal the influence of knowledge graph information on relation extraction tasks.
BIOS: An Algorithmically Generated Biomedical Knowledge Graph
Mar 18, 2022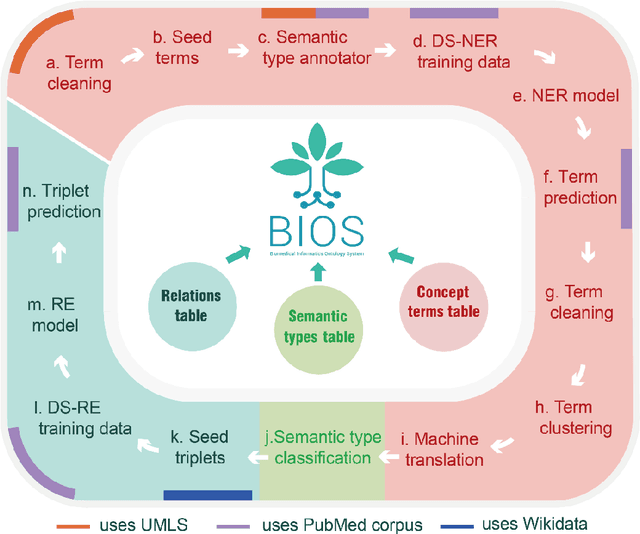
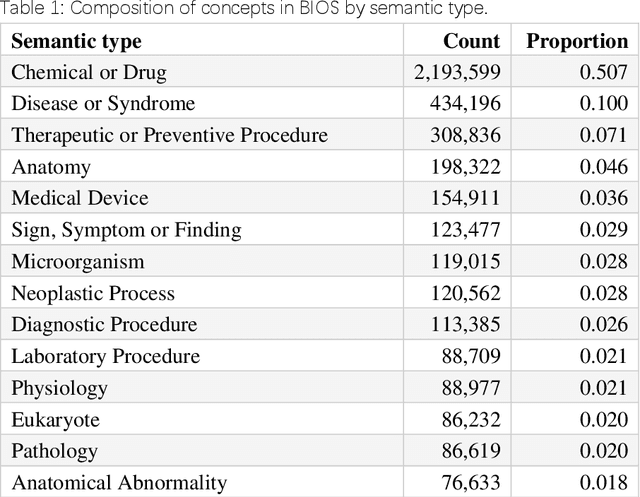
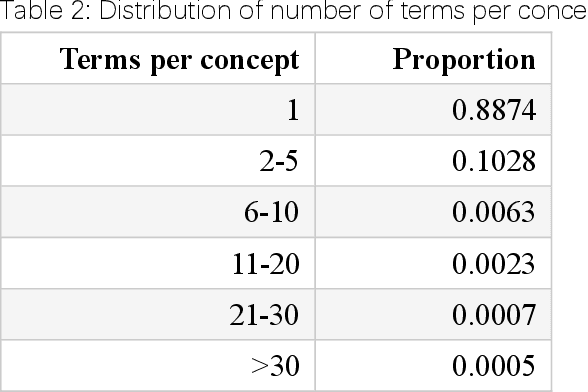

Abstract:Biomedical knowledge graphs (BioMedKGs) are essential infrastructures for biomedical and healthcare big data and artificial intelligence (AI), facilitating natural language processing, model development, and data exchange. For many decades, these knowledge graphs have been built via expert curation, which can no longer catch up with the speed of today's AI development, and a transition to algorithmically generated BioMedKGs is necessary. In this work, we introduce the Biomedical Informatics Ontology System (BIOS), the first large scale publicly available BioMedKG that is fully generated by machine learning algorithms. BIOS currently contains 4.1 million concepts, 7.4 million terms in two languages, and 7.3 million relation triplets. We introduce the methodology for developing BIOS, which covers curation of raw biomedical terms, computationally identifying synonymous terms and aggregating them to create concept nodes, semantic type classification of the concepts, relation identification, and biomedical machine translation. We provide statistics about the current content of BIOS and perform preliminary assessment for term quality, synonym grouping, and relation extraction. Results suggest that machine learning-based BioMedKG development is a totally viable solution for replacing traditional expert curation.
Multimodal Learning on Graphs for Disease Relation Extraction
Mar 16, 2022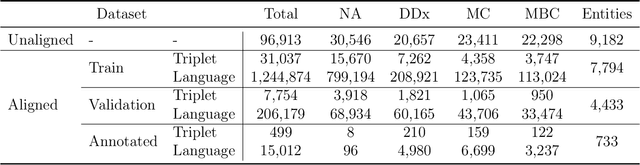

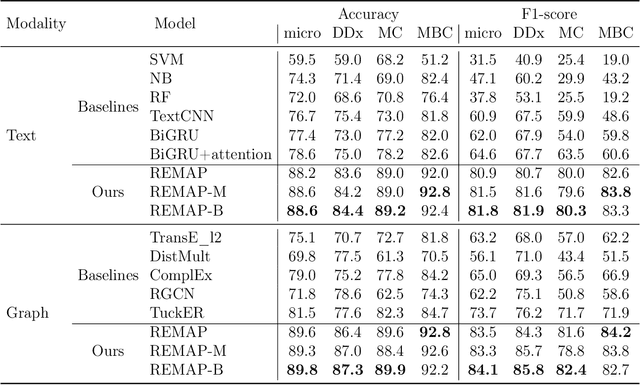
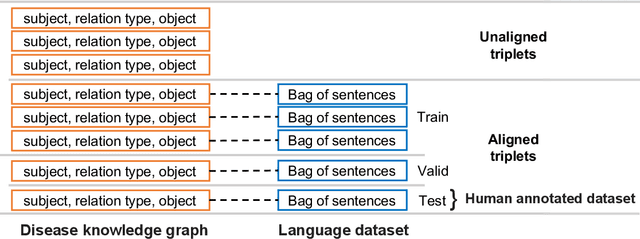
Abstract:Objective: Disease knowledge graphs are a way to connect, organize, and access disparate information about diseases with numerous benefits for artificial intelligence (AI). To create knowledge graphs, it is necessary to extract knowledge from multimodal datasets in the form of relationships between disease concepts and normalize both concepts and relationship types. Methods: We introduce REMAP, a multimodal approach for disease relation extraction and classification. The REMAP machine learning approach jointly embeds a partial, incomplete knowledge graph and a medical language dataset into a compact latent vector space, followed by aligning the multimodal embeddings for optimal disease relation extraction. Results: We apply REMAP approach to a disease knowledge graph with 96,913 relations and a text dataset of 1.24 million sentences. On a dataset annotated by human experts, REMAP improves text-based disease relation extraction by 10.0% (accuracy) and 17.2% (F1-score) by fusing disease knowledge graphs with text information. Further, REMAP leverages text information to recommend new relationships in the knowledge graph, outperforming graph-based methods by 8.4% (accuracy) and 10.4% (F1-score). Discussion: Systematized knowledge is becoming the backbone of AI, creating opportunities to inject semantics into AI and fully integrate it into machine learning algorithms. While prior semantic knowledge can assist in extracting disease relationships from text, existing methods can not fully leverage multimodal datasets. Conclusion: REMAP is a multimodal approach for extracting and classifying disease relationships by fusing structured knowledge and text information. REMAP provides a flexible neural architecture to easily find, access, and validate AI-driven relationships between disease concepts.
High-throughput relation extraction algorithm development associating knowledge articles and electronic health records
Sep 08, 2020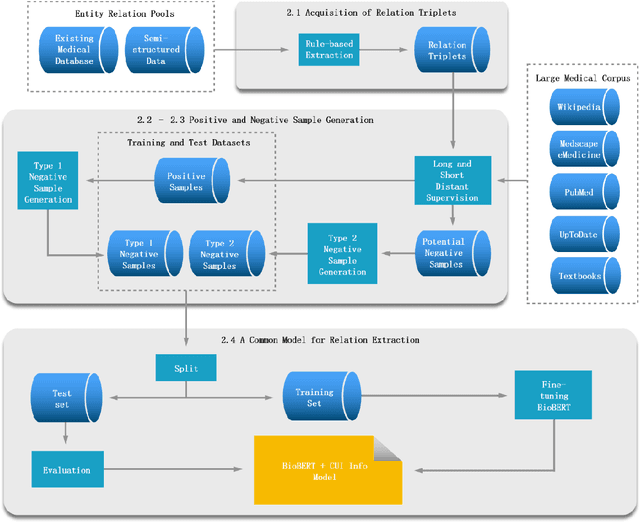
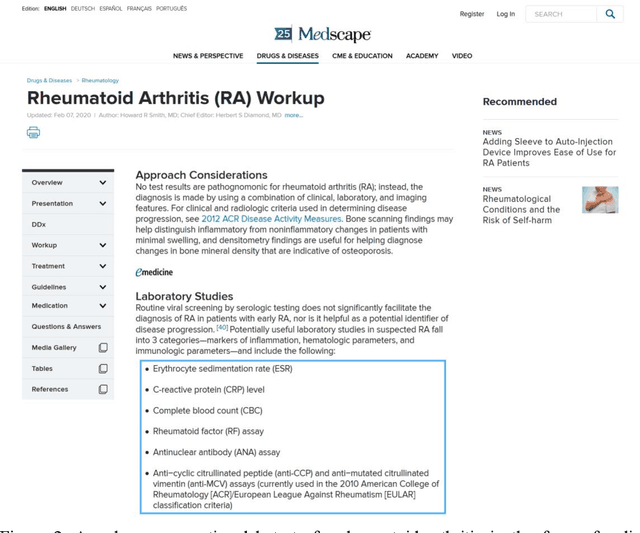
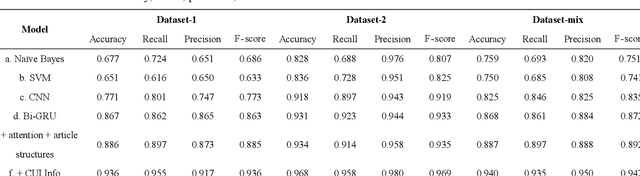
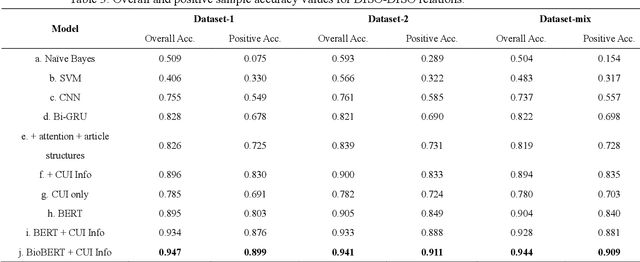
Abstract:Objective: Medical relations are the core components of medical knowledge graphs that are needed for healthcare artificial intelligence. However, the requirement of expert annotation by conventional algorithm development processes creates a major bottleneck for mining new relations. In this paper, we present Hi-RES, a framework for high-throughput relation extraction algorithm development. We also show that combining knowledge articles with electronic health records (EHRs) significantly increases the classification accuracy. Methods: We use relation triplets obtained from structured databases and semistructured webpages to label sentences from target corpora as positive training samples. Two methods are also provided for creating improved negative samples by combining positive samples with na\"ive negative samples. We propose a common model that summarizes sentence information using large-scale pretrained language models and multi-instance attention, which then joins with the concept embeddings trained from the EHRs for relation prediction. Results: We apply the Hi-RES framework to develop classification algorithms for disorder-disorder relations and disorder-location relations. Millions of sentences are created as training data. Using pretrained language models and EHR-based embeddings individually provides considerable accuracy increases over those of previous models. Joining them together further tremendously increases the accuracy to 0.947 and 0.998 for the two sets of relations, respectively, which are 10-17 percentage points higher than those of previous models. Conclusion: Hi-RES is an efficient framework for achieving high-throughput and accurate relation extraction algorithm development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge