Shengxuan Luo
BIOS: An Algorithmically Generated Biomedical Knowledge Graph
Mar 18, 2022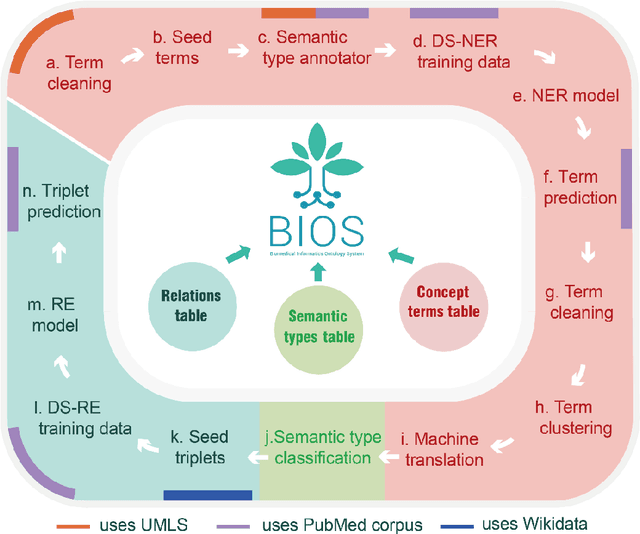
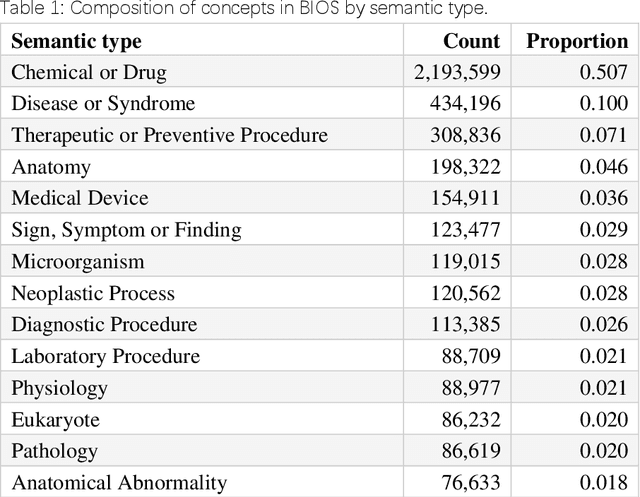
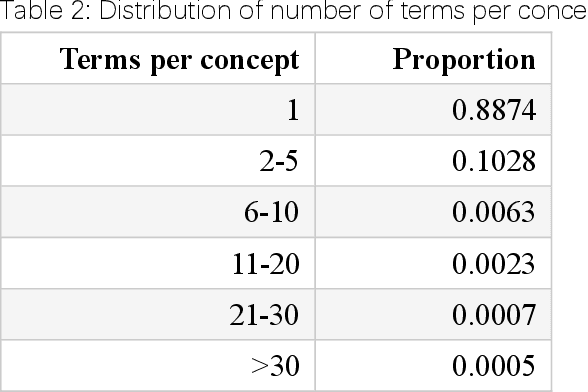

Abstract:Biomedical knowledge graphs (BioMedKGs) are essential infrastructures for biomedical and healthcare big data and artificial intelligence (AI), facilitating natural language processing, model development, and data exchange. For many decades, these knowledge graphs have been built via expert curation, which can no longer catch up with the speed of today's AI development, and a transition to algorithmically generated BioMedKGs is necessary. In this work, we introduce the Biomedical Informatics Ontology System (BIOS), the first large scale publicly available BioMedKG that is fully generated by machine learning algorithms. BIOS currently contains 4.1 million concepts, 7.4 million terms in two languages, and 7.3 million relation triplets. We introduce the methodology for developing BIOS, which covers curation of raw biomedical terms, computationally identifying synonymous terms and aggregating them to create concept nodes, semantic type classification of the concepts, relation identification, and biomedical machine translation. We provide statistics about the current content of BIOS and perform preliminary assessment for term quality, synonym grouping, and relation extraction. Results suggest that machine learning-based BioMedKG development is a totally viable solution for replacing traditional expert curation.
Semi-constraint Optimal Transport for Entity Alignment with Dangling Cases
Mar 11, 2022
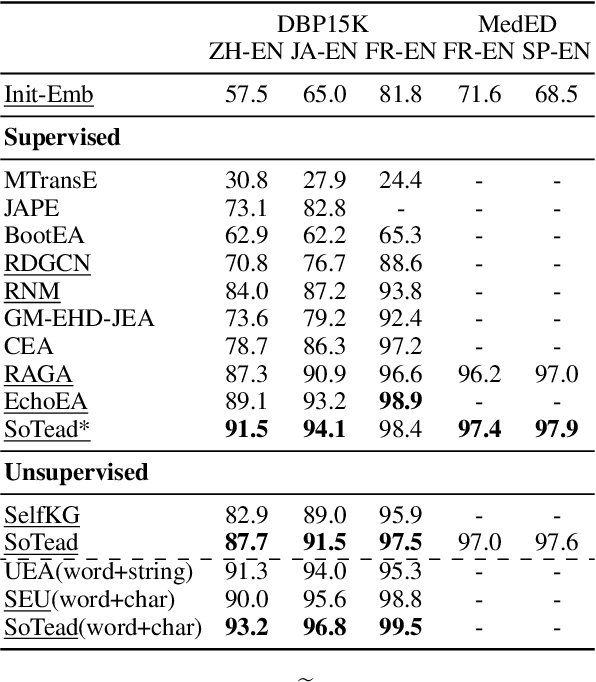

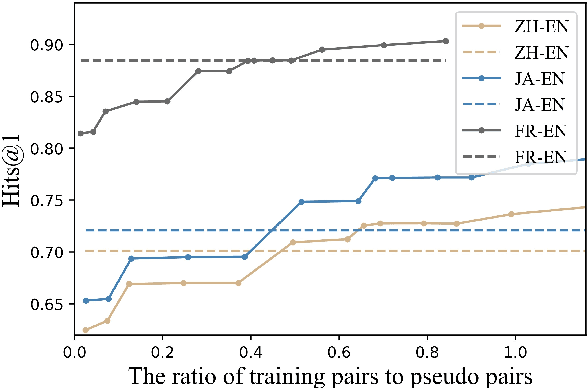
Abstract:Entity alignment (EA) merges knowledge graphs (KGs) by identifying the equivalent entities in different graphs, which can effectively enrich knowledge representations of KGs. However, in practice, different KGs often include dangling entities whose counterparts cannot be found in the other graph, which limits the performance of EA methods. To improve EA with dangling entities, we propose an unsupervised method called Semi-constraint Optimal Transport for Entity Alignment in Dangling cases (SoTead). Our main idea is to model the entity alignment between two KGs as an optimal transport problem from one KG's entities to the others. First, we set pseudo entity pairs between KGs based on pretrained word embeddings. Then, we conduct contrastive metric learning to obtain the transport cost between each entity pair. Finally, we introduce a virtual entity for each KG to "align" the dangling entities from the other KGs, which relaxes the optimization constraints and leads to a semi-constraint optimal transport. In the experimental part, we first show the superiority of SoTead on a commonly-used entity alignment dataset. Besides, to analyze the ability for dangling entity detection with other baselines, we construct a medical cross-lingual knowledge graph dataset, MedED, where our SoTead also reaches state-of-the-art performance.
An Accurate Unsupervised Method for Joint Entity Alignment and Dangling Entity Detection
Mar 10, 2022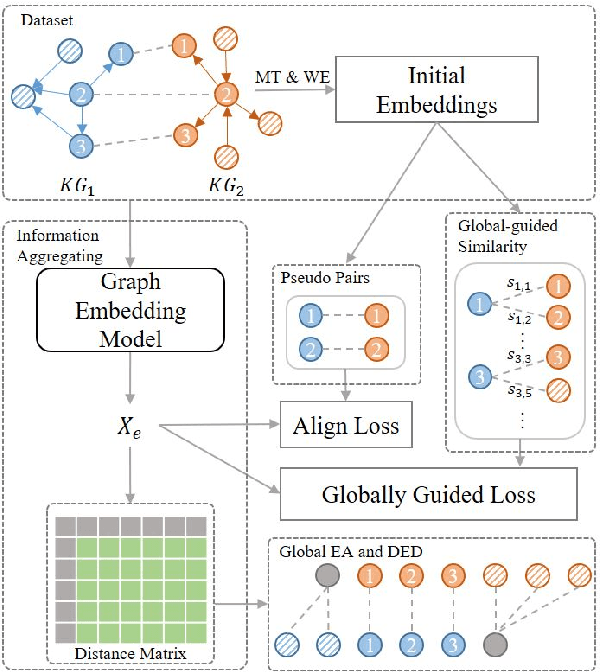
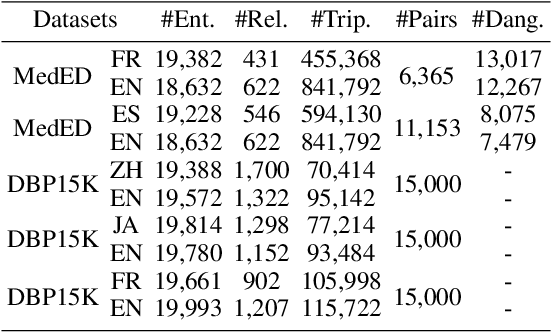
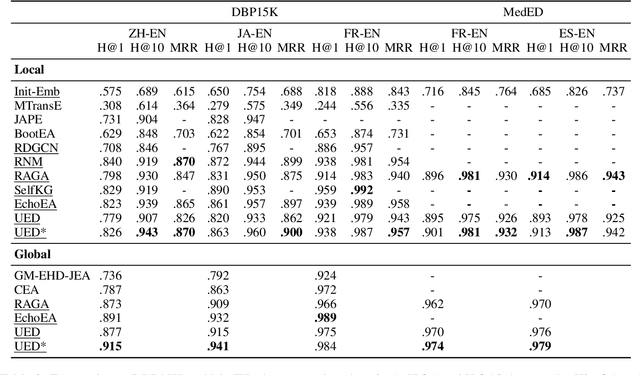
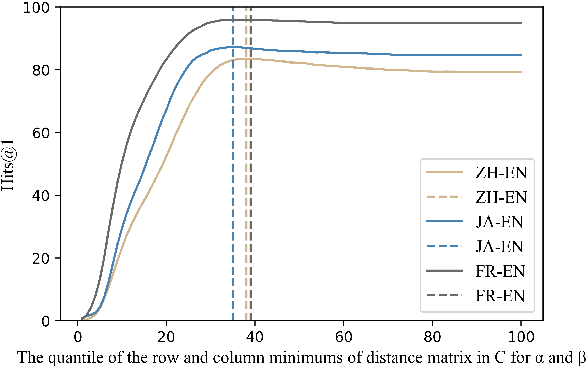
Abstract:Knowledge graph integration typically suffers from the widely existing dangling entities that cannot find alignment cross knowledge graphs (KGs). The dangling entity set is unavailable in most real-world scenarios, and manually mining the entity pairs that consist of entities with the same meaning is labor-consuming. In this paper, we propose a novel accurate Unsupervised method for joint Entity alignment (EA) and Dangling entity detection (DED), called UED. The UED mines the literal semantic information to generate pseudo entity pairs and globally guided alignment information for EA and then utilizes the EA results to assist the DED. We construct a medical cross-lingual knowledge graph dataset, MedED, providing data for both the EA and DED tasks. Extensive experiments demonstrate that in the EA task, UED achieves EA results comparable to those of state-of-the-art supervised EA baselines and outperforms the current state-of-the-art EA methods by combining supervised EA data. For the DED task, UED obtains high-quality results without supervision.
Sentence Alignment with Parallel Documents Helps Biomedical Machine Translation
Apr 17, 2021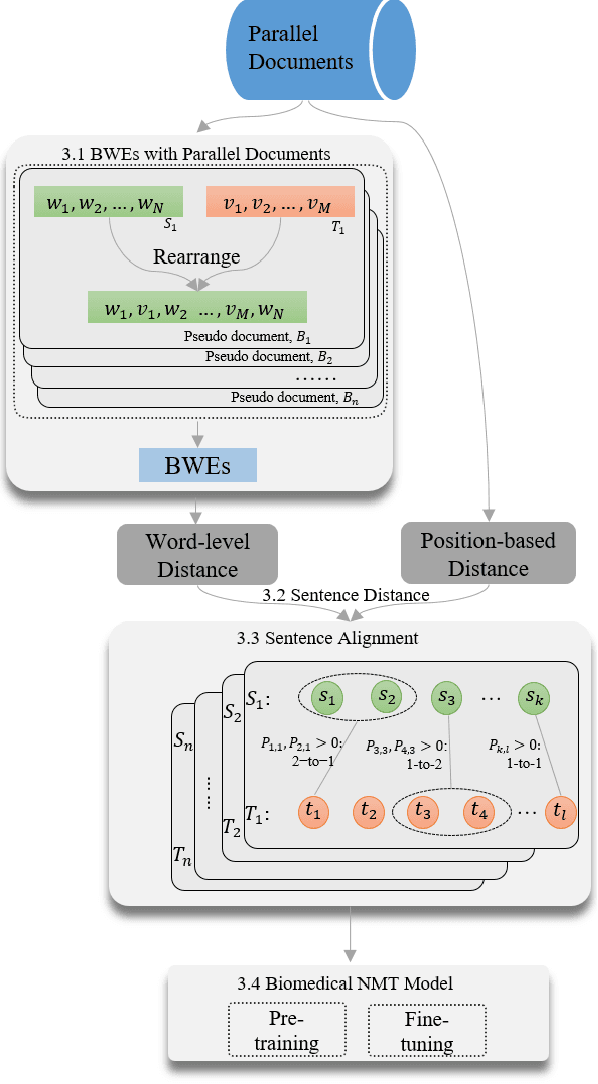
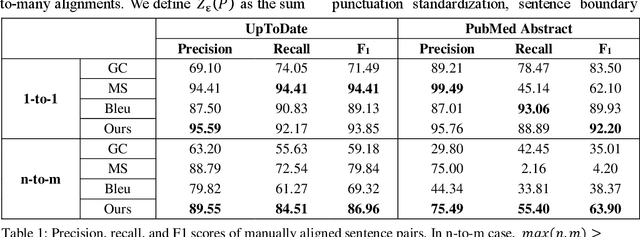
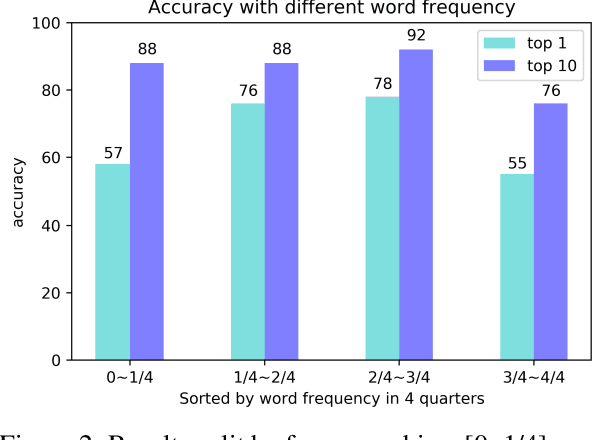
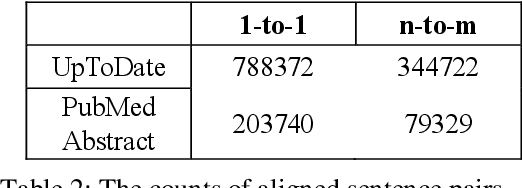
Abstract:The existing neural machine translation system has achieved near human-level performance in general domain in some languages, but the lack of parallel corpora poses a key problem in specific domains. In biomedical domain, the parallel corpus is less accessible. This work presents a new unsupervised sentence alignment method and explores features in training biomedical neural machine translation (NMT) systems. We use a simple but effective way to build bilingual word embeddings (BWEs) to evaluate bilingual word similarity and transferred the sentence alignment problem into an extended earth mover's distance (EMD) problem. The proposed method achieved high accuracy in both 1-to-1 and many-to-many cases. Pre-training in general domain, the larger in-domain dataset and n-to-m sentence pairs benefit the NMT model. Fine-tuning in domain corpus helps the translation model learns more terminology and fits the in-domain style of text.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge