Huaiyuan Ying
Seed-Prover: Deep and Broad Reasoning for Automated Theorem Proving
Aug 01, 2025Abstract:LLMs have demonstrated strong mathematical reasoning abilities by leveraging reinforcement learning with long chain-of-thought, yet they continue to struggle with theorem proving due to the lack of clear supervision signals when solely using natural language. Dedicated domain-specific languages like Lean provide clear supervision via formal verification of proofs, enabling effective training through reinforcement learning. In this work, we propose \textbf{Seed-Prover}, a lemma-style whole-proof reasoning model. Seed-Prover can iteratively refine its proof based on Lean feedback, proved lemmas, and self-summarization. To solve IMO-level contest problems, we design three test-time inference strategies that enable both deep and broad reasoning. Seed-Prover proves $78.1\%$ of formalized past IMO problems, saturates MiniF2F, and achieves over 50\% on PutnamBench, outperforming the previous state-of-the-art by a large margin. To address the lack of geometry support in Lean, we introduce a geometry reasoning engine \textbf{Seed-Geometry}, which outperforms previous formal geometry engines. We use these two systems to participate in IMO 2025 and fully prove 5 out of 6 problems. This work represents a significant advancement in automated mathematical reasoning, demonstrating the effectiveness of formal verification with long chain-of-thought reasoning.
DR.EHR: Dense Retrieval for Electronic Health Record with Knowledge Injection and Synthetic Data
Jul 24, 2025Abstract:Electronic Health Records (EHRs) are pivotal in clinical practices, yet their retrieval remains a challenge mainly due to semantic gap issues. Recent advancements in dense retrieval offer promising solutions but existing models, both general-domain and biomedical-domain, fall short due to insufficient medical knowledge or mismatched training corpora. This paper introduces \texttt{DR.EHR}, a series of dense retrieval models specifically tailored for EHR retrieval. We propose a two-stage training pipeline utilizing MIMIC-IV discharge summaries to address the need for extensive medical knowledge and large-scale training data. The first stage involves medical entity extraction and knowledge injection from a biomedical knowledge graph, while the second stage employs large language models to generate diverse training data. We train two variants of \texttt{DR.EHR}, with 110M and 7B parameters, respectively. Evaluated on the CliniQ benchmark, our models significantly outperforms all existing dense retrievers, achieving state-of-the-art results. Detailed analyses confirm our models' superiority across various match and query types, particularly in challenging semantic matches like implication and abbreviation. Ablation studies validate the effectiveness of each pipeline component, and supplementary experiments on EHR QA datasets demonstrate the models' generalizability on natural language questions, including complex ones with multiple entities. This work significantly advances EHR retrieval, offering a robust solution for clinical applications.
Evaluating Entity Retrieval in Electronic Health Records: a Semantic Gap Perspective
Feb 10, 2025



Abstract:Entity retrieval plays a crucial role in the utilization of Electronic Health Records (EHRs) and is applied across a wide range of clinical practices. However, a comprehensive evaluation of this task is lacking due to the absence of a public benchmark. In this paper, we propose the development and release of a novel benchmark for evaluating entity retrieval in EHRs, with a particular focus on the semantic gap issue. Using discharge summaries from the MIMIC-III dataset, we incorporate ICD codes and prescription labels associated with the notes as queries, and annotate relevance judgments using GPT-4. In total, we use 1,000 patient notes, generate 1,246 queries, and provide over 77,000 relevance annotations. To offer the first assessment of the semantic gap, we introduce a novel classification system for relevance matches. Leveraging GPT-4, we categorize each relevant pair into one of five categories: string, synonym, abbreviation, hyponym, and implication. Using the proposed benchmark, we evaluate several retrieval methods, including BM25, query expansion, and state-of-the-art dense retrievers. Our findings show that BM25 provides a strong baseline but struggles with semantic matches. Query expansion significantly improves performance, though it slightly reduces string match capabilities. Dense retrievers outperform traditional methods, particularly for semantic matches, and general-domain dense retrievers often surpass those trained specifically in the biomedical domain.
GENIE: Generative Note Information Extraction model for structuring EHR data
Jan 30, 2025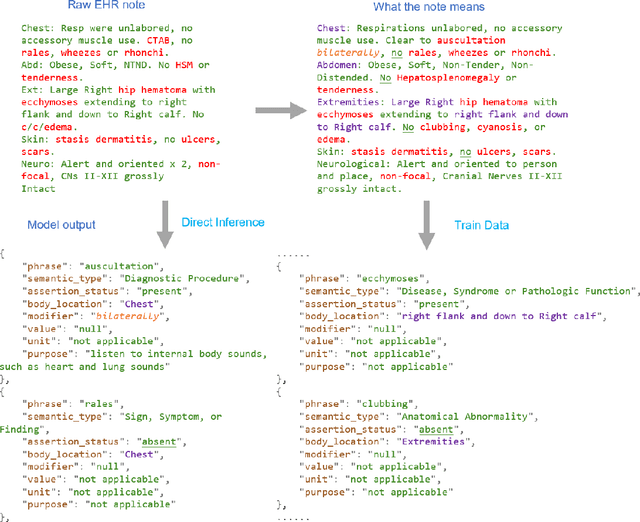
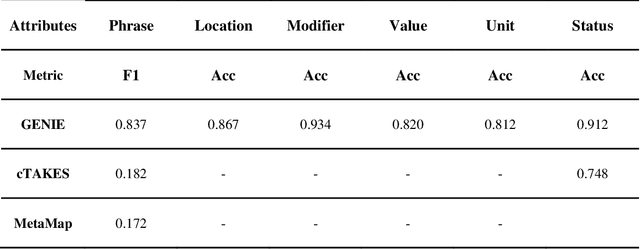
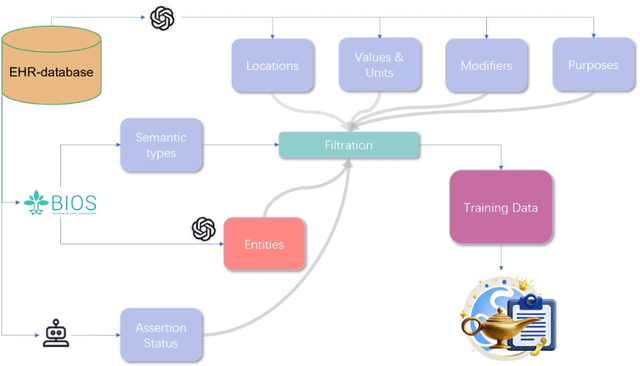
Abstract:Electronic Health Records (EHRs) hold immense potential for advancing healthcare, offering rich, longitudinal data that combines structured information with valuable insights from unstructured clinical notes. However, the unstructured nature of clinical text poses significant challenges for secondary applications. Traditional methods for structuring EHR free-text data, such as rule-based systems and multi-stage pipelines, are often limited by their time-consuming configurations and inability to adapt across clinical notes from diverse healthcare settings. Few systems provide a comprehensive attribute extraction for terminologies. While giant large language models (LLMs) like GPT-4 and LLaMA 405B excel at structuring tasks, they are slow, costly, and impractical for large-scale use. To overcome these limitations, we introduce GENIE, a Generative Note Information Extraction system that leverages LLMs to streamline the structuring of unstructured clinical text into usable data with standardized format. GENIE processes entire paragraphs in a single pass, extracting entities, assertion statuses, locations, modifiers, values, and purposes with high accuracy. Its unified, end-to-end approach simplifies workflows, reduces errors, and eliminates the need for extensive manual intervention. Using a robust data preparation pipeline and fine-tuned small scale LLMs, GENIE achieves competitive performance across multiple information extraction tasks, outperforming traditional tools like cTAKES and MetaMap and can handle extra attributes to be extracted. GENIE strongly enhances real-world applicability and scalability in healthcare systems. By open-sourcing the model and test data, we aim to encourage collaboration and drive further advancements in EHR structurization.
InternLM2.5-StepProver: Advancing Automated Theorem Proving via Expert Iteration on Large-Scale LEAN Problems
Oct 21, 2024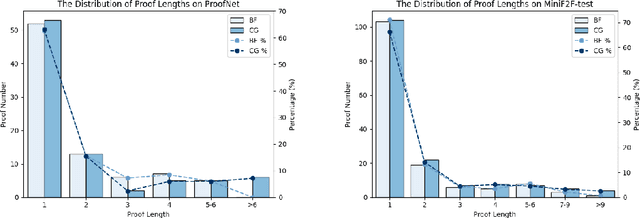

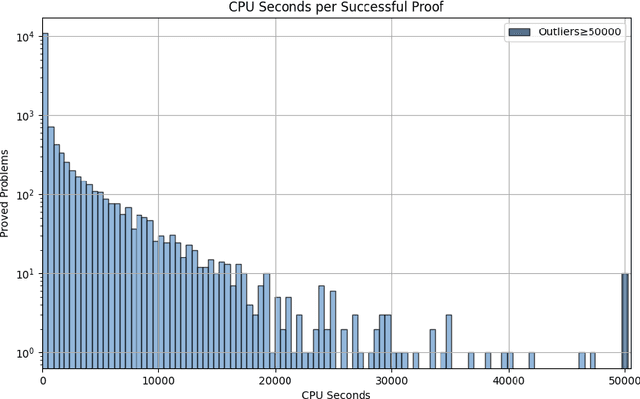

Abstract:Large Language Models (LLMs) have emerged as powerful tools in mathematical theorem proving, particularly when utilizing formal languages such as LEAN. The major learning paradigm is expert iteration, which necessitates a pre-defined dataset comprising numerous mathematical problems. In this process, LLMs attempt to prove problems within the dataset and iteratively refine their capabilities through self-training on the proofs they discover. We propose to use large scale LEAN problem datasets Lean-workbook for expert iteration with more than 20,000 CPU days. During expert iteration, we found log-linear trends between solved problem amount with proof length and CPU usage. We train a critic model to select relatively easy problems for policy models to make trials and guide the model to search for deeper proofs. InternLM2.5-StepProver achieves open-source state-of-the-art on MiniF2F, Lean-Workbook-Plus, ProofNet, and Putnam benchmarks. Specifically, it achieves a pass of 65.9% on the MiniF2F-test and proves (or disproves) 17.0% of problems in Lean-Workbook-Plus which shows a significant improvement compared to only 9.5% of problems proved when Lean-Workbook-Plus was released. We open-source our models and searched proofs at https://github.com/InternLM/InternLM-Math and https://huggingface.co/datasets/internlm/Lean-Workbook.
Lean Workbook: A large-scale Lean problem set formalized from natural language math problems
Jun 07, 2024Abstract:Large language models have demonstrated impressive capabilities across various natural language processing tasks, especially in solving mathematical problems. However, large language models are not good at math theorem proving using formal languages like Lean. A significant challenge in this area is the scarcity of training data available in these formal languages. To address this issue, we propose a novel pipeline that iteratively generates and filters synthetic data to translate natural language mathematical problems into Lean 4 statements, and vice versa. Our results indicate that the synthetic data pipeline can provide useful training data and improve the performance of LLMs in translating and understanding complex mathematical problems and proofs. Our final dataset contains about 57K formal-informal question pairs along with searched proof from the math contest forum and 21 new IMO questions. We open-source our code at https://github.com/InternLM/InternLM-Math and our data at https://huggingface.co/datasets/InternLM/Lean-Workbook.
InternLM2 Technical Report
Mar 26, 2024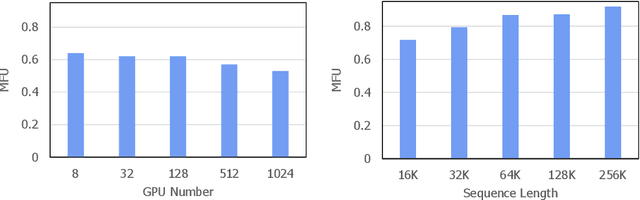
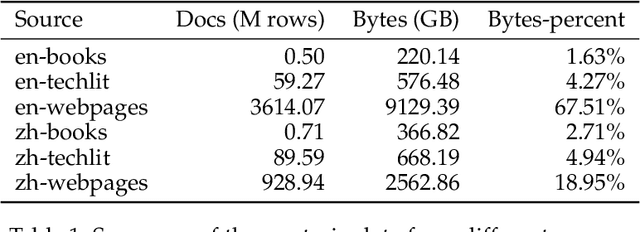

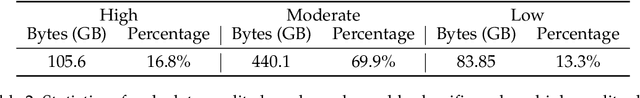
Abstract:The evolution of Large Language Models (LLMs) like ChatGPT and GPT-4 has sparked discussions on the advent of Artificial General Intelligence (AGI). However, replicating such advancements in open-source models has been challenging. This paper introduces InternLM2, an open-source LLM that outperforms its predecessors in comprehensive evaluations across 6 dimensions and 30 benchmarks, long-context modeling, and open-ended subjective evaluations through innovative pre-training and optimization techniques. The pre-training process of InternLM2 is meticulously detailed, highlighting the preparation of diverse data types including text, code, and long-context data. InternLM2 efficiently captures long-term dependencies, initially trained on 4k tokens before advancing to 32k tokens in pre-training and fine-tuning stages, exhibiting remarkable performance on the 200k ``Needle-in-a-Haystack" test. InternLM2 is further aligned using Supervised Fine-Tuning (SFT) and a novel Conditional Online Reinforcement Learning from Human Feedback (COOL RLHF) strategy that addresses conflicting human preferences and reward hacking. By releasing InternLM2 models in different training stages and model sizes, we provide the community with insights into the model's evolution.
A Question Answering Based Pipeline for Comprehensive Chinese EHR Information Extraction
Feb 17, 2024Abstract:Electronic health records (EHRs) hold significant value for research and applications. As a new way of information extraction, question answering (QA) can extract more flexible information than conventional methods and is more accessible to clinical researchers, but its progress is impeded by the scarcity of annotated data. In this paper, we propose a novel approach that automatically generates training data for transfer learning of QA models. Our pipeline incorporates a preprocessing module to handle challenges posed by extraction types that are not readily compatible with extractive QA frameworks, including cases with discontinuous answers and many-to-one relationships. The obtained QA model exhibits excellent performance on subtasks of information extraction in EHRs, and it can effectively handle few-shot or zero-shot settings involving yes-no questions. Case studies and ablation studies demonstrate the necessity of each component in our design, and the resulting model is deemed suitable for practical use.
InternLM-Math: Open Math Large Language Models Toward Verifiable Reasoning
Feb 09, 2024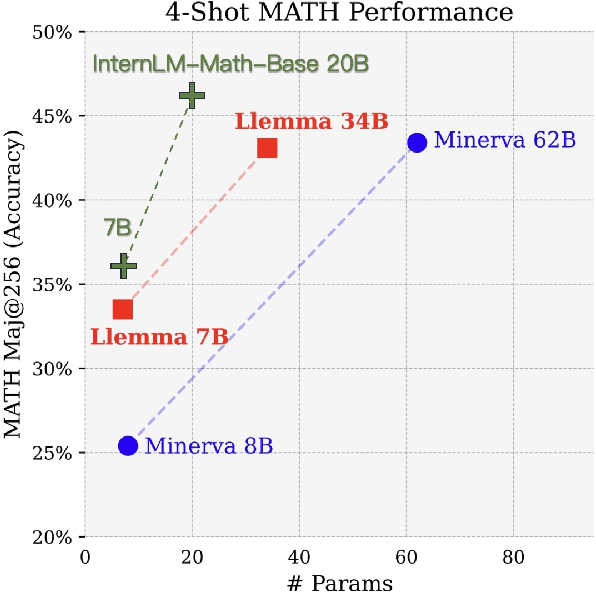
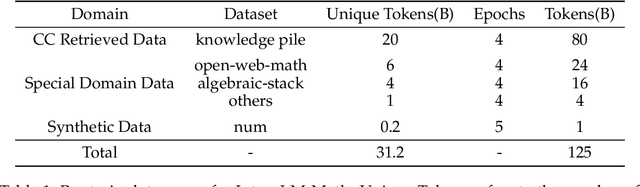
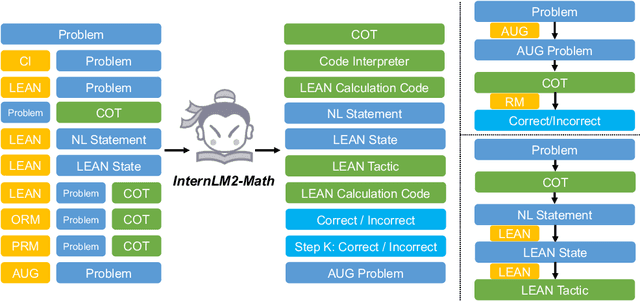
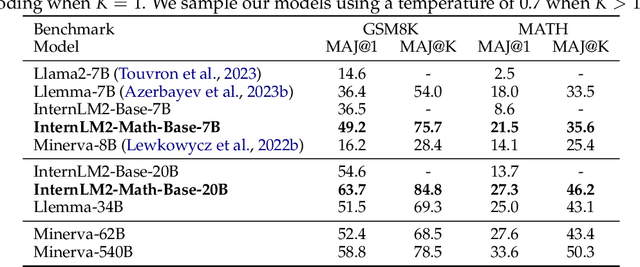
Abstract:The math abilities of large language models can represent their abstract reasoning ability. In this paper, we introduce and open-source our math reasoning LLMs InternLM-Math which is continue pre-trained from InternLM2. We unify chain-of-thought reasoning, reward modeling, formal reasoning, data augmentation, and code interpreter in a unified seq2seq format and supervise our model to be a versatile math reasoner, verifier, prover, and augmenter. These abilities can be used to develop the next math LLMs or self-iteration. InternLM-Math obtains open-sourced state-of-the-art performance under the setting of in-context learning, supervised fine-tuning, and code-assisted reasoning in various informal and formal benchmarks including GSM8K, MATH, Hungary math exam, MathBench-ZH, and MiniF2F. Our pre-trained model achieves 30.3 on the MiniF2F test set without fine-tuning. We further explore how to use LEAN to solve math problems and study its performance under the setting of multi-task learning which shows the possibility of using LEAN as a unified platform for solving and proving in math. Our models, codes, and data are released at \url{https://github.com/InternLM/InternLM-Math}.
CoRTEx: Contrastive Learning for Representing Terms via Explanations with Applications on Constructing Biomedical Knowledge Graphs
Dec 13, 2023

Abstract:Objective: Biomedical Knowledge Graphs play a pivotal role in various biomedical research domains. Concurrently, term clustering emerges as a crucial step in constructing these knowledge graphs, aiming to identify synonymous terms. Due to a lack of knowledge, previous contrastive learning models trained with Unified Medical Language System (UMLS) synonyms struggle at clustering difficult terms and do not generalize well beyond UMLS terms. In this work, we leverage the world knowledge from Large Language Models (LLMs) and propose Contrastive Learning for Representing Terms via Explanations (CoRTEx) to enhance term representation and significantly improves term clustering. Materials and Methods: The model training involves generating explanations for a cleaned subset of UMLS terms using ChatGPT. We employ contrastive learning, considering term and explanation embeddings simultaneously, and progressively introduce hard negative samples. Additionally, a ChatGPT-assisted BIRCH algorithm is designed for efficient clustering of a new ontology. Results: We established a clustering test set and a hard negative test set, where our model consistently achieves the highest F1 score. With CoRTEx embeddings and the modified BIRCH algorithm, we grouped 35,580,932 terms from the Biomedical Informatics Ontology System (BIOS) into 22,104,559 clusters with O(N) queries to ChatGPT. Case studies highlight the model's efficacy in handling challenging samples, aided by information from explanations. Conclusion: By aligning terms to their explanations, CoRTEx demonstrates superior accuracy over benchmark models and robustness beyond its training set, and it is suitable for clustering terms for large-scale biomedical ontologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge