Sheng Yu
LLM-Aligned Geographic Item Tokenization for Local-Life Recommendation
Nov 18, 2025Abstract:Recent advances in Large Language Models (LLMs) have enhanced text-based recommendation by enriching traditional ID-based methods with semantic generalization capabilities. Text-based methods typically encode item textual information via prompt design and generate discrete semantic IDs through item tokenization. However, in domain-specific tasks such as local-life services, simply injecting location information into prompts fails to capture fine-grained spatial characteristics and real-world distance awareness among items. To address this, we propose LGSID, an LLM-Aligned Geographic Item Tokenization Framework for Local-life Recommendation. This framework consists of two key components: (1) RL-based Geographic LLM Alignment, and (2) Hierarchical Geographic Item Tokenization. In the RL-based alignment module, we initially train a list-wise reward model to capture real-world spatial relationships among items. We then introduce a novel G-DPO algorithm that uses pre-trained reward model to inject generalized spatial knowledge and collaborative signals into LLMs while preserving their semantic understanding. Furthermore, we propose a hierarchical geographic item tokenization strategy, where primary tokens are derived from discrete spatial and content attributes, and residual tokens are refined using the aligned LLM's geographic representation vectors. Extensive experiments on real-world Kuaishou industry datasets show that LGSID consistently outperforms state-of-the-art discriminative and generative recommendation models. Ablation studies, visualizations, and case studies further validate its effectiveness.
A Plug-and-Play Spatially-Constrained Representation Enhancement Framework for Local-Life Recommendation
Nov 17, 2025Abstract:Local-life recommendation have witnessed rapid growth, providing users with convenient access to daily essentials. However, this domain faces two key challenges: (1) spatial constraints, driven by the requirements of the local-life scenario, where items are usually shown only to users within a limited geographic area, indirectly reducing their exposure probability; and (2) long-tail sparsity, where few popular items dominate user interactions, while many high-quality long-tail items are largely overlooked due to imbalanced interaction opportunities. Existing methods typically adopt a user-centric perspective, such as modeling spatial user preferences or enhancing long-tail representations with collaborative filtering signals. However, we argue that an item-centric perspective is more suitable for this domain, focusing on enhancing long-tail items representation that align with the spatially-constrained characteristics of local lifestyle services. To tackle this issue, we propose ReST, a Plug-And-Play Spatially-Constrained Representation Enhancement Framework for Long-Tail Local-Life Recommendation. Specifically, we first introduce a Meta ID Warm-up Network, which initializes fundamental ID representations by injecting their basic attribute-level semantic information. Subsequently, we propose a novel Spatially-Constrained ID Representation Enhancement Network (SIDENet) based on contrastive learning, which incorporates two efficient strategies: a spatially-constrained hard sampling strategy and a dynamic representation alignment strategy. This design adaptively identifies weak ID representations based on their attribute-level information during training. It additionally enhances them by capturing latent item relationships within the spatially-constrained characteristics of local lifestyle services, while preserving compatibility with popular items.
DR.EHR: Dense Retrieval for Electronic Health Record with Knowledge Injection and Synthetic Data
Jul 24, 2025Abstract:Electronic Health Records (EHRs) are pivotal in clinical practices, yet their retrieval remains a challenge mainly due to semantic gap issues. Recent advancements in dense retrieval offer promising solutions but existing models, both general-domain and biomedical-domain, fall short due to insufficient medical knowledge or mismatched training corpora. This paper introduces \texttt{DR.EHR}, a series of dense retrieval models specifically tailored for EHR retrieval. We propose a two-stage training pipeline utilizing MIMIC-IV discharge summaries to address the need for extensive medical knowledge and large-scale training data. The first stage involves medical entity extraction and knowledge injection from a biomedical knowledge graph, while the second stage employs large language models to generate diverse training data. We train two variants of \texttt{DR.EHR}, with 110M and 7B parameters, respectively. Evaluated on the CliniQ benchmark, our models significantly outperforms all existing dense retrievers, achieving state-of-the-art results. Detailed analyses confirm our models' superiority across various match and query types, particularly in challenging semantic matches like implication and abbreviation. Ablation studies validate the effectiveness of each pipeline component, and supplementary experiments on EHR QA datasets demonstrate the models' generalizability on natural language questions, including complex ones with multiple entities. This work significantly advances EHR retrieval, offering a robust solution for clinical applications.
Evaluating Entity Retrieval in Electronic Health Records: a Semantic Gap Perspective
Feb 10, 2025



Abstract:Entity retrieval plays a crucial role in the utilization of Electronic Health Records (EHRs) and is applied across a wide range of clinical practices. However, a comprehensive evaluation of this task is lacking due to the absence of a public benchmark. In this paper, we propose the development and release of a novel benchmark for evaluating entity retrieval in EHRs, with a particular focus on the semantic gap issue. Using discharge summaries from the MIMIC-III dataset, we incorporate ICD codes and prescription labels associated with the notes as queries, and annotate relevance judgments using GPT-4. In total, we use 1,000 patient notes, generate 1,246 queries, and provide over 77,000 relevance annotations. To offer the first assessment of the semantic gap, we introduce a novel classification system for relevance matches. Leveraging GPT-4, we categorize each relevant pair into one of five categories: string, synonym, abbreviation, hyponym, and implication. Using the proposed benchmark, we evaluate several retrieval methods, including BM25, query expansion, and state-of-the-art dense retrievers. Our findings show that BM25 provides a strong baseline but struggles with semantic matches. Query expansion significantly improves performance, though it slightly reduces string match capabilities. Dense retrievers outperform traditional methods, particularly for semantic matches, and general-domain dense retrievers often surpass those trained specifically in the biomedical domain.
GENIE: Generative Note Information Extraction model for structuring EHR data
Jan 30, 2025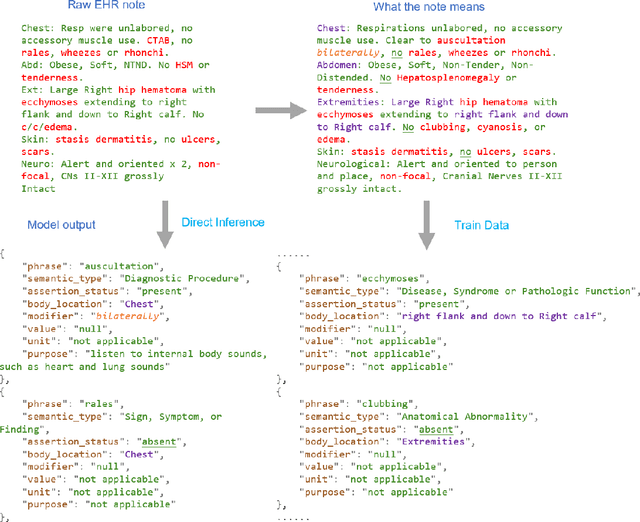
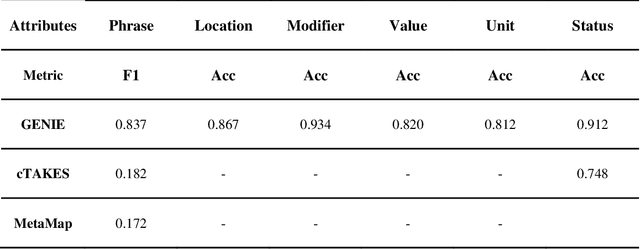
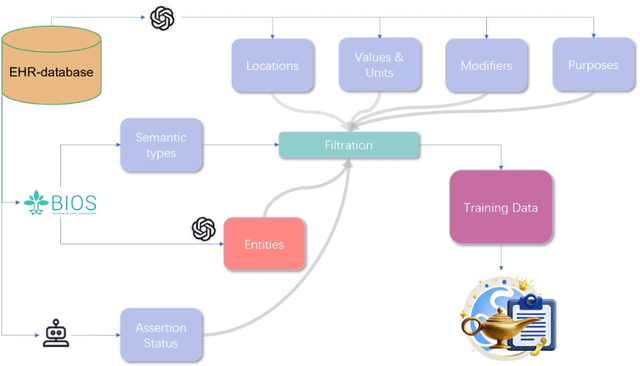
Abstract:Electronic Health Records (EHRs) hold immense potential for advancing healthcare, offering rich, longitudinal data that combines structured information with valuable insights from unstructured clinical notes. However, the unstructured nature of clinical text poses significant challenges for secondary applications. Traditional methods for structuring EHR free-text data, such as rule-based systems and multi-stage pipelines, are often limited by their time-consuming configurations and inability to adapt across clinical notes from diverse healthcare settings. Few systems provide a comprehensive attribute extraction for terminologies. While giant large language models (LLMs) like GPT-4 and LLaMA 405B excel at structuring tasks, they are slow, costly, and impractical for large-scale use. To overcome these limitations, we introduce GENIE, a Generative Note Information Extraction system that leverages LLMs to streamline the structuring of unstructured clinical text into usable data with standardized format. GENIE processes entire paragraphs in a single pass, extracting entities, assertion statuses, locations, modifiers, values, and purposes with high accuracy. Its unified, end-to-end approach simplifies workflows, reduces errors, and eliminates the need for extensive manual intervention. Using a robust data preparation pipeline and fine-tuned small scale LLMs, GENIE achieves competitive performance across multiple information extraction tasks, outperforming traditional tools like cTAKES and MetaMap and can handle extra attributes to be extracted. GENIE strongly enhances real-world applicability and scalability in healthcare systems. By open-sourcing the model and test data, we aim to encourage collaboration and drive further advancements in EHR structurization.
Black-Box Segmentation of Electronic Medical Records
Sep 29, 2024
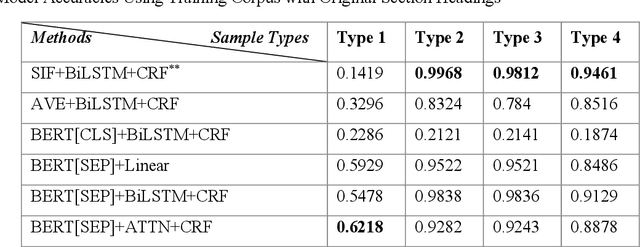
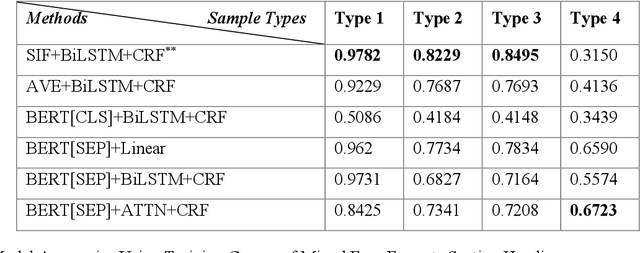
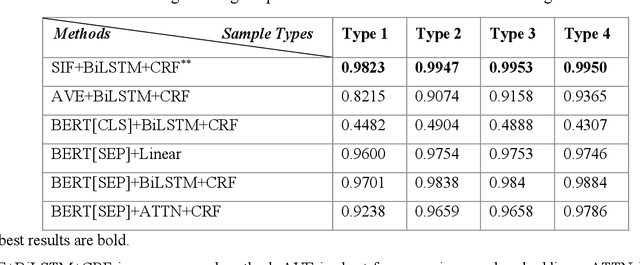
Abstract:Electronic medical records (EMRs) contain the majority of patients' healthcare details. It is an abundant resource for developing an automatic healthcare system. Most of the natural language processing (NLP) studies on EMR processing, such as concept extraction, are adversely affected by the inaccurate segmentation of EMR sections. At the same time, not enough attention has been given to the accurate sectioning of EMRs. The information that may occur in section structures is unvalued. This work focuses on the segmentation of EMRs and proposes a black-box segmentation method using a simple sentence embedding model and neural network, along with a proper training method. To achieve universal adaptivity, we train our model on the dataset with different section headings formats. We compare several advanced deep learning-based NLP methods, and our method achieves the best segmentation accuracies (above 98%) on various test data with a proper training corpus.
A Question Answering Based Pipeline for Comprehensive Chinese EHR Information Extraction
Feb 17, 2024Abstract:Electronic health records (EHRs) hold significant value for research and applications. As a new way of information extraction, question answering (QA) can extract more flexible information than conventional methods and is more accessible to clinical researchers, but its progress is impeded by the scarcity of annotated data. In this paper, we propose a novel approach that automatically generates training data for transfer learning of QA models. Our pipeline incorporates a preprocessing module to handle challenges posed by extraction types that are not readily compatible with extractive QA frameworks, including cases with discontinuous answers and many-to-one relationships. The obtained QA model exhibits excellent performance on subtasks of information extraction in EHRs, and it can effectively handle few-shot or zero-shot settings involving yes-no questions. Case studies and ablation studies demonstrate the necessity of each component in our design, and the resulting model is deemed suitable for practical use.
CAREForMe: Contextual Multi-Armed Bandit Recommendation Framework for Mental Health
Jan 26, 2024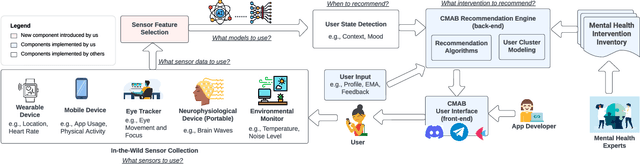
Abstract:The COVID-19 pandemic has intensified the urgency for effective and accessible mental health interventions in people's daily lives. Mobile Health (mHealth) solutions, such as AI Chatbots and Mindfulness Apps, have gained traction as they expand beyond traditional clinical settings to support daily life. However, the effectiveness of current mHealth solutions is impeded by the lack of context-awareness, personalization, and modularity to foster their reusability. This paper introduces CAREForMe, a contextual multi-armed bandit (CMAB) recommendation framework for mental health. Designed with context-awareness, personalization, and modularity at its core, CAREForMe harnesses mobile sensing and integrates online learning algorithms with user clustering capability to deliver timely, personalized recommendations. With its modular design, CAREForMe serves as both a customizable recommendation framework to guide future research, and a collaborative platform to facilitate interdisciplinary contributions in mHealth research. We showcase CAREForMe's versatility through its implementation across various platforms (e.g., Discord, Telegram) and its customization to diverse recommendation features.
High-throughput Biomedical Relation Extraction for Semi-Structured Web Articles Empowered by Large Language Models
Dec 15, 2023Abstract:Objective: To develop a high-throughput biomedical relation extraction system that takes advantage of the large language models' (LLMs) reading comprehension ability and biomedical world knowledge in a scalable and evidential manner. Methods: We formulate the relation extraction task as a simple binary classification problem for large language models such as ChatGPT. Specifically, LLMs make the decision based on the external corpus and its world knowledge, giving the reason for the judgment to factual verification. This method is tailored for semi-structured web articles, wherein we designate the main title as the tail entity and explicitly incorporate it into the context, and the potential head entities are matched based on a biomedical thesaurus. Moreover, lengthy contents are sliced into text chunks, embedded, and retrieved with additional embedding models, ensuring compatibility with the context window size constraints of available open-source LLMs. Results: Using an open-source LLM, we extracted 304315 relation triplets of three distinct relation types from four reputable biomedical websites. To assess the efficacy of the basic pipeline employed for biomedical relation extraction, we curated a benchmark dataset annotated by a medical expert. Evaluation results indicate that the pipeline exhibits performance comparable to that of GPT-4. Case studies further illuminate challenges faced by contemporary LLMs in the context of biomedical relation extraction for semi-structured web articles. Conclusion: The proposed method has demonstrated its effectiveness in leveraging the strengths of LLMs for high-throughput biomedical relation extraction. Its adaptability is evident, as it can be seamlessly extended to diverse semi-structured biomedical websites, facilitating the extraction of various types of biomedical relations with ease.
CoRTEx: Contrastive Learning for Representing Terms via Explanations with Applications on Constructing Biomedical Knowledge Graphs
Dec 13, 2023

Abstract:Objective: Biomedical Knowledge Graphs play a pivotal role in various biomedical research domains. Concurrently, term clustering emerges as a crucial step in constructing these knowledge graphs, aiming to identify synonymous terms. Due to a lack of knowledge, previous contrastive learning models trained with Unified Medical Language System (UMLS) synonyms struggle at clustering difficult terms and do not generalize well beyond UMLS terms. In this work, we leverage the world knowledge from Large Language Models (LLMs) and propose Contrastive Learning for Representing Terms via Explanations (CoRTEx) to enhance term representation and significantly improves term clustering. Materials and Methods: The model training involves generating explanations for a cleaned subset of UMLS terms using ChatGPT. We employ contrastive learning, considering term and explanation embeddings simultaneously, and progressively introduce hard negative samples. Additionally, a ChatGPT-assisted BIRCH algorithm is designed for efficient clustering of a new ontology. Results: We established a clustering test set and a hard negative test set, where our model consistently achieves the highest F1 score. With CoRTEx embeddings and the modified BIRCH algorithm, we grouped 35,580,932 terms from the Biomedical Informatics Ontology System (BIOS) into 22,104,559 clusters with O(N) queries to ChatGPT. Case studies highlight the model's efficacy in handling challenging samples, aided by information from explanations. Conclusion: By aligning terms to their explanations, CoRTEx demonstrates superior accuracy over benchmark models and robustness beyond its training set, and it is suitable for clustering terms for large-scale biomedical ontologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge