Hongyi Yuan
Visual Generation Unlocks Human-Like Reasoning through Multimodal World Models
Jan 27, 2026Abstract:Humans construct internal world models and reason by manipulating the concepts within these models. Recent advances in AI, particularly chain-of-thought (CoT) reasoning, approximate such human cognitive abilities, where world models are believed to be embedded within large language models. Expert-level performance in formal and abstract domains such as mathematics and programming has been achieved in current systems by relying predominantly on verbal reasoning. However, they still lag far behind humans in domains like physical and spatial intelligence, which require richer representations and prior knowledge. The emergence of unified multimodal models (UMMs) capable of both verbal and visual generation has therefore sparked interest in more human-like reasoning grounded in complementary multimodal pathways, though their benefits remain unclear. From a world-model perspective, this paper presents the first principled study of when and how visual generation benefits reasoning. Our key position is the visual superiority hypothesis: for certain tasks--particularly those grounded in the physical world--visual generation more naturally serves as world models, whereas purely verbal world models encounter bottlenecks arising from representational limitations or insufficient prior knowledge. Theoretically, we formalize internal world modeling as a core component of CoT reasoning and analyze distinctions among different forms of world models. Empirically, we identify tasks that necessitate interleaved visual-verbal CoT reasoning, constructing a new evaluation suite, VisWorld-Eval. Controlled experiments on a state-of-the-art UMM show that interleaved CoT significantly outperforms purely verbal CoT on tasks that favor visual world modeling, but offers no clear advantage otherwise. Together, this work clarifies the potential of multimodal world modeling for more powerful, human-like multimodal AI.
Seed-Prover: Deep and Broad Reasoning for Automated Theorem Proving
Aug 01, 2025Abstract:LLMs have demonstrated strong mathematical reasoning abilities by leveraging reinforcement learning with long chain-of-thought, yet they continue to struggle with theorem proving due to the lack of clear supervision signals when solely using natural language. Dedicated domain-specific languages like Lean provide clear supervision via formal verification of proofs, enabling effective training through reinforcement learning. In this work, we propose \textbf{Seed-Prover}, a lemma-style whole-proof reasoning model. Seed-Prover can iteratively refine its proof based on Lean feedback, proved lemmas, and self-summarization. To solve IMO-level contest problems, we design three test-time inference strategies that enable both deep and broad reasoning. Seed-Prover proves $78.1\%$ of formalized past IMO problems, saturates MiniF2F, and achieves over 50\% on PutnamBench, outperforming the previous state-of-the-art by a large margin. To address the lack of geometry support in Lean, we introduce a geometry reasoning engine \textbf{Seed-Geometry}, which outperforms previous formal geometry engines. We use these two systems to participate in IMO 2025 and fully prove 5 out of 6 problems. This work represents a significant advancement in automated mathematical reasoning, demonstrating the effectiveness of formal verification with long chain-of-thought reasoning.
Evaluating Entity Retrieval in Electronic Health Records: a Semantic Gap Perspective
Feb 10, 2025



Abstract:Entity retrieval plays a crucial role in the utilization of Electronic Health Records (EHRs) and is applied across a wide range of clinical practices. However, a comprehensive evaluation of this task is lacking due to the absence of a public benchmark. In this paper, we propose the development and release of a novel benchmark for evaluating entity retrieval in EHRs, with a particular focus on the semantic gap issue. Using discharge summaries from the MIMIC-III dataset, we incorporate ICD codes and prescription labels associated with the notes as queries, and annotate relevance judgments using GPT-4. In total, we use 1,000 patient notes, generate 1,246 queries, and provide over 77,000 relevance annotations. To offer the first assessment of the semantic gap, we introduce a novel classification system for relevance matches. Leveraging GPT-4, we categorize each relevant pair into one of five categories: string, synonym, abbreviation, hyponym, and implication. Using the proposed benchmark, we evaluate several retrieval methods, including BM25, query expansion, and state-of-the-art dense retrievers. Our findings show that BM25 provides a strong baseline but struggles with semantic matches. Query expansion significantly improves performance, though it slightly reduces string match capabilities. Dense retrievers outperform traditional methods, particularly for semantic matches, and general-domain dense retrievers often surpass those trained specifically in the biomedical domain.
GENIE: Generative Note Information Extraction model for structuring EHR data
Jan 30, 2025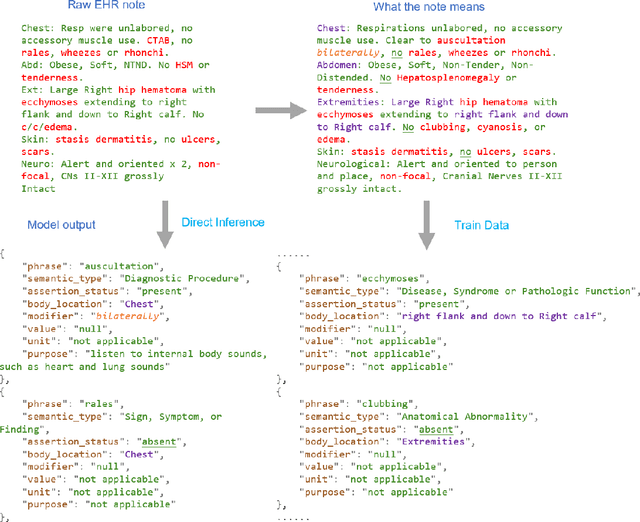
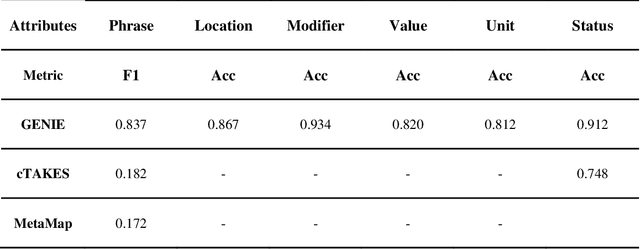
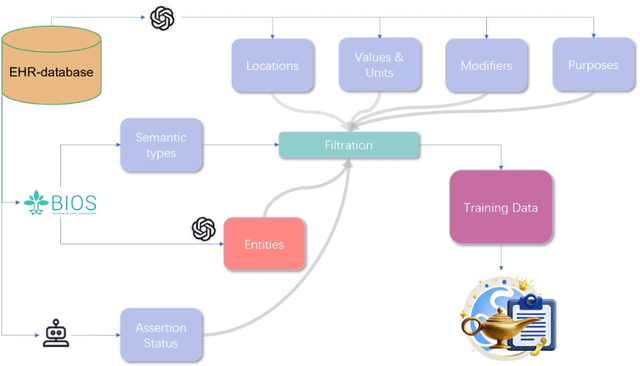
Abstract:Electronic Health Records (EHRs) hold immense potential for advancing healthcare, offering rich, longitudinal data that combines structured information with valuable insights from unstructured clinical notes. However, the unstructured nature of clinical text poses significant challenges for secondary applications. Traditional methods for structuring EHR free-text data, such as rule-based systems and multi-stage pipelines, are often limited by their time-consuming configurations and inability to adapt across clinical notes from diverse healthcare settings. Few systems provide a comprehensive attribute extraction for terminologies. While giant large language models (LLMs) like GPT-4 and LLaMA 405B excel at structuring tasks, they are slow, costly, and impractical for large-scale use. To overcome these limitations, we introduce GENIE, a Generative Note Information Extraction system that leverages LLMs to streamline the structuring of unstructured clinical text into usable data with standardized format. GENIE processes entire paragraphs in a single pass, extracting entities, assertion statuses, locations, modifiers, values, and purposes with high accuracy. Its unified, end-to-end approach simplifies workflows, reduces errors, and eliminates the need for extensive manual intervention. Using a robust data preparation pipeline and fine-tuned small scale LLMs, GENIE achieves competitive performance across multiple information extraction tasks, outperforming traditional tools like cTAKES and MetaMap and can handle extra attributes to be extracted. GENIE strongly enhances real-world applicability and scalability in healthcare systems. By open-sourcing the model and test data, we aim to encourage collaboration and drive further advancements in EHR structurization.
Unified Representation of Genomic and Biomedical Concepts through Multi-Task, Multi-Source Contrastive Learning
Oct 14, 2024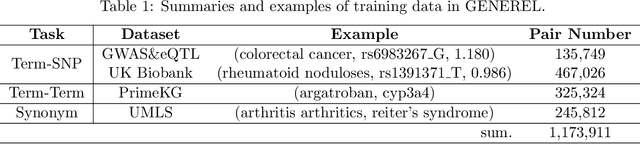
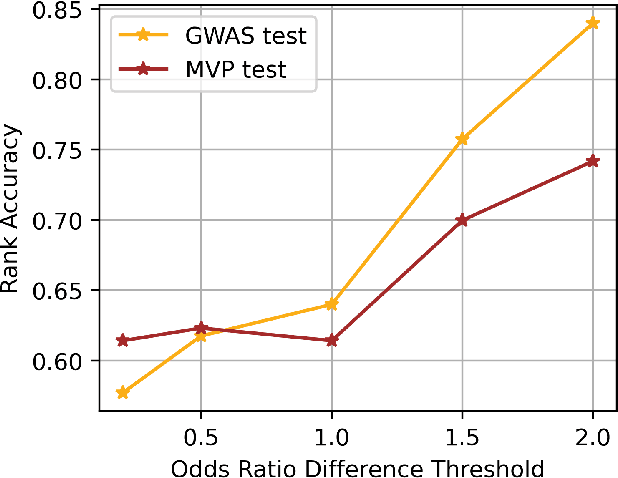
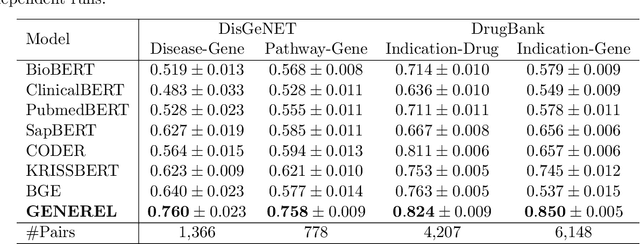
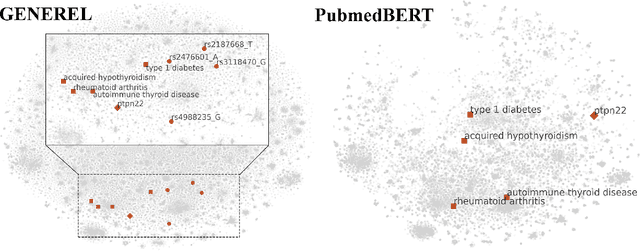
Abstract:We introduce GENomic Encoding REpresentation with Language Model (GENEREL), a framework designed to bridge genetic and biomedical knowledge bases. What sets GENEREL apart is its ability to fine-tune language models to infuse biological knowledge behind clinical concepts such as diseases and medications. This fine-tuning enables the model to capture complex biomedical relationships more effectively, enriching the understanding of how genomic data connects to clinical outcomes. By constructing a unified embedding space for biomedical concepts and a wide range of common SNPs from sources such as patient-level data, biomedical knowledge graphs, and GWAS summaries, GENEREL aligns the embeddings of SNPs and clinical concepts through multi-task contrastive learning. This allows the model to adapt to diverse natural language representations of biomedical concepts while bypassing the limitations of traditional code mapping systems across different data sources. Our experiments demonstrate GENEREL's ability to effectively capture the nuanced relationships between SNPs and clinical concepts. GENEREL also emerges to discern the degree of relatedness, potentially allowing for a more refined identification of concepts. This pioneering approach in constructing a unified embedding system for both SNPs and biomedical concepts enhances the potential for data integration and discovery in biomedical research.
Black-Box Segmentation of Electronic Medical Records
Sep 29, 2024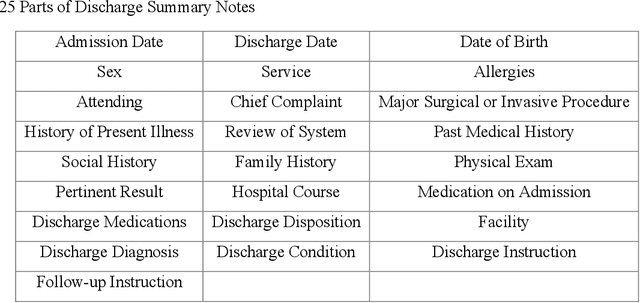
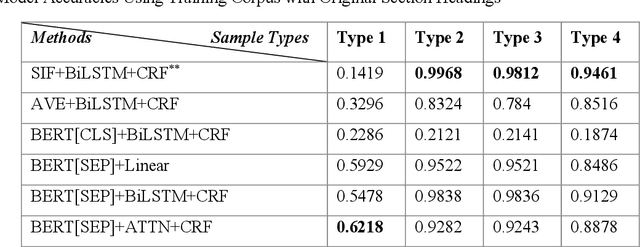
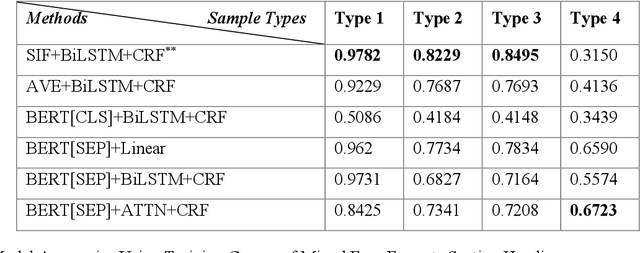
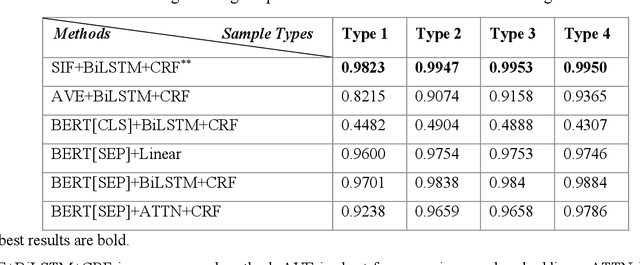
Abstract:Electronic medical records (EMRs) contain the majority of patients' healthcare details. It is an abundant resource for developing an automatic healthcare system. Most of the natural language processing (NLP) studies on EMR processing, such as concept extraction, are adversely affected by the inaccurate segmentation of EMR sections. At the same time, not enough attention has been given to the accurate sectioning of EMRs. The information that may occur in section structures is unvalued. This work focuses on the segmentation of EMRs and proposes a black-box segmentation method using a simple sentence embedding model and neural network, along with a proper training method. To achieve universal adaptivity, we train our model on the dataset with different section headings formats. We compare several advanced deep learning-based NLP methods, and our method achieves the best segmentation accuracies (above 98%) on various test data with a proper training corpus.
Routing to the Expert: Efficient Reward-guided Ensemble of Large Language Models
Nov 15, 2023Abstract:The complementary potential of Large Language Models (LLM) assumes off-the-shelf LLMs have heterogeneous expertise in a wide range of domains and tasks so that an ensemble of LLMs can achieve consistently better performance. Existing ensemble methods for LLMs mainly focus on reward model ranking of outputs, leading to significant computation overhead. To combat this issue, we revisit the complementary potential of LLMs and further elaborate it by mining latent expertise with off-the-shelf reward models. We propose Zooter, a reward-guided routing method distilling rewards on training queries to train a routing function, which can precisely distribute each query to the LLM with expertise about it. We also integrate a tag-based label enhancement to mitigate noise from uncertainty when using rewards as silver supervision. Zooter shows computation efficiency in inference as it introduces only a minor computation overhead of a routing function compared with reward model ranking methods. We evaluate Zooter on a comprehensive benchmark collection with 26 subsets on different domains and tasks. Zooter outperforms the best single model on average and ranks first on 44% of tasks, even surpassing multiple reward model ranking methods.
Speculative Contrastive Decoding
Nov 15, 2023


Abstract:Large language models (LLMs) have shown extraordinary performance in various language tasks, but high computational requirements hinder their widespread deployment. Speculative decoding, which uses amateur models to predict the generation of expert models, has been proposed as a way to accelerate LLM inference. However, speculative decoding focuses on acceleration instead of making the best use of the token distribution from amateur models. We proposed Speculative Contrastive Decoding (SCD), an accelerated decoding method leveraging the natural contrast between expert and amateur models in speculative decoding. Comprehensive evaluations on four benchmarks show that SCD can achieve similar acceleration factors as speculative decoding while further improving the generation quality as the contrastive decoding. The analysis of token probabilities further demonstrates the compatibility between speculative and contrastive decoding. Overall, SCD provides an effective approach to enhance the decoding quality of LLMs while saving computational resources.
How Abilities in Large Language Models are Affected by Supervised Fine-tuning Data Composition
Oct 09, 2023



Abstract:Large language models (LLMs) with enormous pre-training tokens and parameter amounts emerge abilities, including math reasoning, code generation, and instruction following. These abilities are further enhanced by supervised fine-tuning (SFT). The open-source community has studied on ad-hoc SFT for each ability, while proprietary LLMs are versatile for all abilities. It is important to investigate how to unlock them with multiple abilities via SFT. In this study, we specifically focus on the data composition between mathematical reasoning, code generation, and general human-aligning abilities during SFT. From a scaling perspective, we investigate the relationship between model abilities and various factors including data amounts, data composition ratio, model parameters, and SFT strategies. Our experiments reveal that different abilities exhibit different scaling patterns, and larger models generally show superior performance with the same amount of data. Mathematical reasoning and code generation improve as data amounts increase consistently, while the general ability is enhanced with about a thousand samples and improves slowly. We find data composition results in various abilities improvements with low data amounts, while conflicts of abilities with high data amounts. Our experiments further show that composition data amount impacts performance, while the influence of composition ratio is insignificant. Regarding the SFT strategies, we evaluate sequential learning multiple abilities are prone to catastrophic forgetting. Our proposed Dual-stage Mixed Fine-tuning (DMT) strategy learns specialized abilities first and then learns general abilities with a small amount of specialized data to prevent forgetting, offering a promising solution to learn multiple abilities with different scaling patterns.
Qwen Technical Report
Sep 28, 2023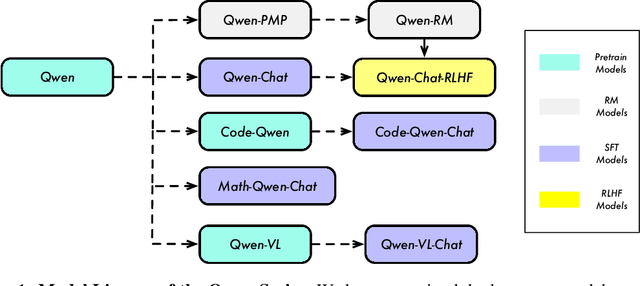
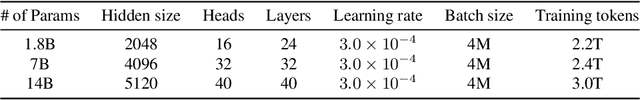
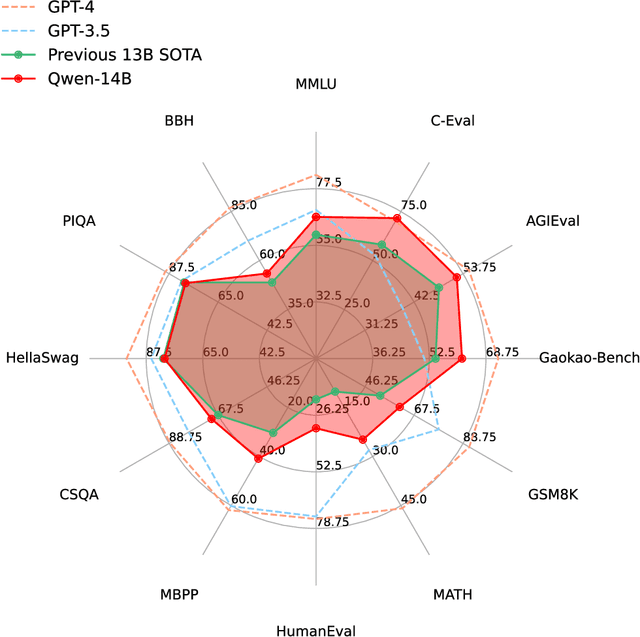
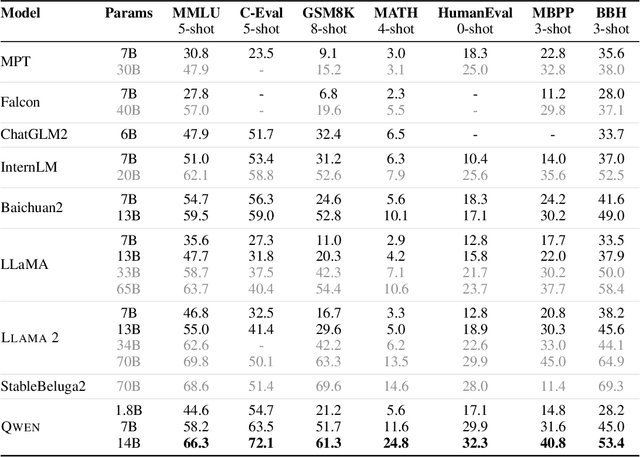
Abstract:Large language models (LLMs) have revolutionized the field of artificial intelligence, enabling natural language processing tasks that were previously thought to be exclusive to humans. In this work, we introduce Qwen, the first installment of our large language model series. Qwen is a comprehensive language model series that encompasses distinct models with varying parameter counts. It includes Qwen, the base pretrained language models, and Qwen-Chat, the chat models finetuned with human alignment techniques. The base language models consistently demonstrate superior performance across a multitude of downstream tasks, and the chat models, particularly those trained using Reinforcement Learning from Human Feedback (RLHF), are highly competitive. The chat models possess advanced tool-use and planning capabilities for creating agent applications, showcasing impressive performance even when compared to bigger models on complex tasks like utilizing a code interpreter. Furthermore, we have developed coding-specialized models, Code-Qwen and Code-Qwen-Chat, as well as mathematics-focused models, Math-Qwen-Chat, which are built upon base language models. These models demonstrate significantly improved performance in comparison with open-source models, and slightly fall behind the proprietary models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge