Alexandre Pereira
Unified Representation of Genomic and Biomedical Concepts through Multi-Task, Multi-Source Contrastive Learning
Oct 14, 2024
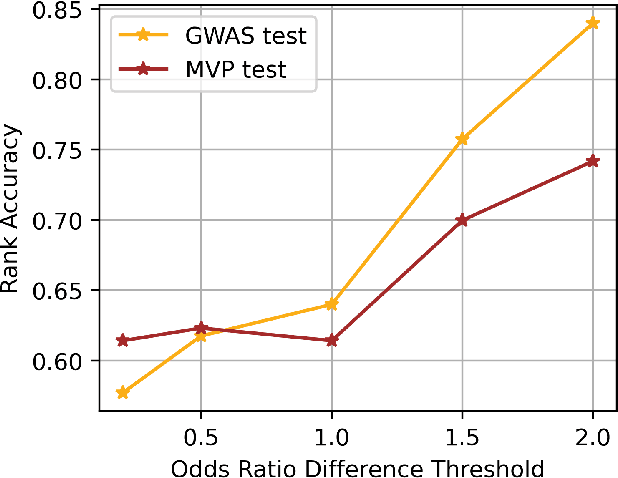
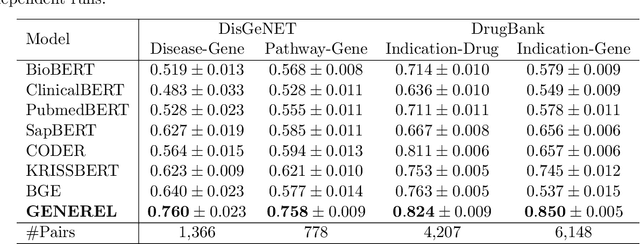
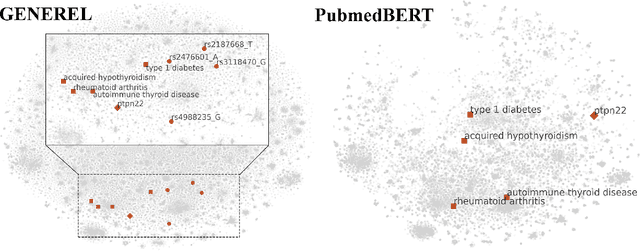
Abstract:We introduce GENomic Encoding REpresentation with Language Model (GENEREL), a framework designed to bridge genetic and biomedical knowledge bases. What sets GENEREL apart is its ability to fine-tune language models to infuse biological knowledge behind clinical concepts such as diseases and medications. This fine-tuning enables the model to capture complex biomedical relationships more effectively, enriching the understanding of how genomic data connects to clinical outcomes. By constructing a unified embedding space for biomedical concepts and a wide range of common SNPs from sources such as patient-level data, biomedical knowledge graphs, and GWAS summaries, GENEREL aligns the embeddings of SNPs and clinical concepts through multi-task contrastive learning. This allows the model to adapt to diverse natural language representations of biomedical concepts while bypassing the limitations of traditional code mapping systems across different data sources. Our experiments demonstrate GENEREL's ability to effectively capture the nuanced relationships between SNPs and clinical concepts. GENEREL also emerges to discern the degree of relatedness, potentially allowing for a more refined identification of concepts. This pioneering approach in constructing a unified embedding system for both SNPs and biomedical concepts enhances the potential for data integration and discovery in biomedical research.
Using Deep Learning for Morphological Classification in Pigs with a Focus on Sanitary Monitoring
Mar 13, 2024Abstract:The aim of this paper is to evaluate the use of D-CNN (Deep Convolutional Neural Networks) algorithms to classify pig body conditions in normal or not normal conditions, with a focus on characteristics that are observed in sanitary monitoring, and were used six different algorithms to do this task. The study focused on five pig characteristics, being these caudophagy, ear hematoma, scratches on the body, redness, and natural stains (brown or black). The results of the study showed that D-CNN was effective in classifying deviations in pig body morphologies related to skin characteristics. The evaluation was conducted by analyzing the performance metrics Precision, Recall, and F-score, as well as the statistical analyses ANOVA and the Scott-Knott test. The contribution of this article is characterized by the proposal of using D-CNN networks for morphological classification in pigs, with a focus on characteristics identified in sanitary monitoring. Among the best results, the average Precision metric of 80.6\% to classify caudophagy was achieved for the InceptionResNetV2 network, indicating the potential use of this technology for the proposed task. Additionally, a new image database was created, containing various pig's distinct body characteristics, which can serve as data for future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge