Katherine Liao
Representation Learning to Advance Multi-institutional Studies with Electronic Health Record Data
Feb 12, 2025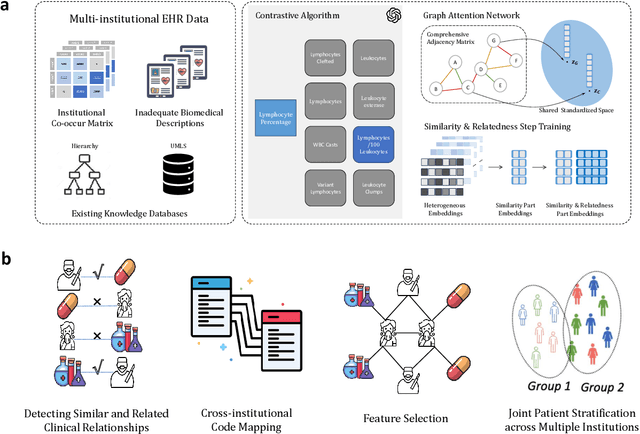
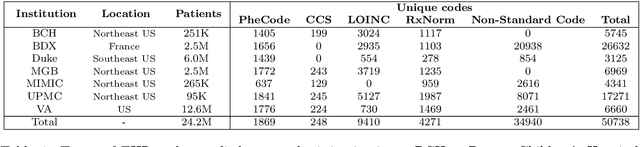
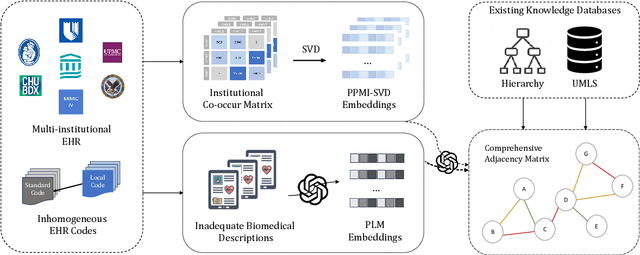
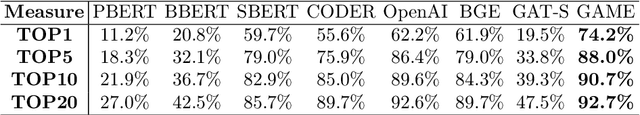
Abstract:The adoption of EHRs has expanded opportunities to leverage data-driven algorithms in clinical care and research. A major bottleneck in effectively conducting multi-institutional EHR studies is the data heterogeneity across systems with numerous codes that either do not exist or represent different clinical concepts across institutions. The need for data privacy further limits the feasibility of including multi-institutional patient-level data required to study similarities and differences across patient subgroups. To address these challenges, we developed the GAME algorithm. Tested and validated across 7 institutions and 2 languages, GAME integrates data in several levels: (1) at the institutional level with knowledge graphs to establish relationships between codes and existing knowledge sources, providing the medical context for standard codes and their relationship to each other; (2) between institutions, leveraging language models to determine the relationships between institution-specific codes with established standard codes; and (3) quantifying the strength of the relationships between codes using a graph attention network. Jointly trained embeddings are created using transfer and federated learning to preserve data privacy. In this study, we demonstrate the applicability of GAME in selecting relevant features as inputs for AI-driven algorithms in a range of conditions, e.g., heart failure, rheumatoid arthritis. We then highlight the application of GAME harmonized multi-institutional EHR data in a study of Alzheimer's disease outcomes and suicide risk among patients with mental health disorders, without sharing patient-level data outside individual institutions.
Unified Representation of Genomic and Biomedical Concepts through Multi-Task, Multi-Source Contrastive Learning
Oct 14, 2024
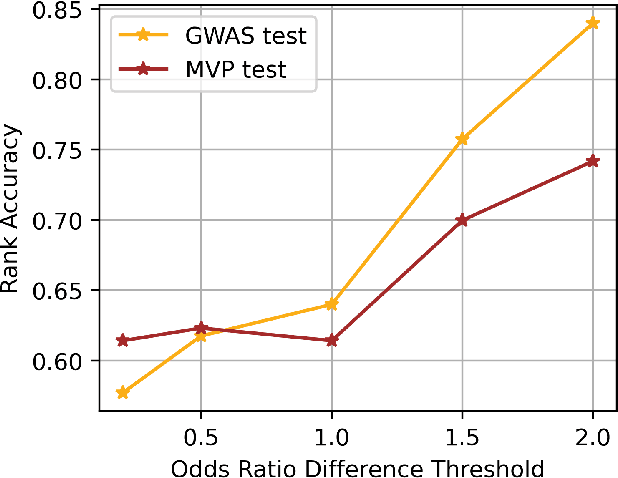
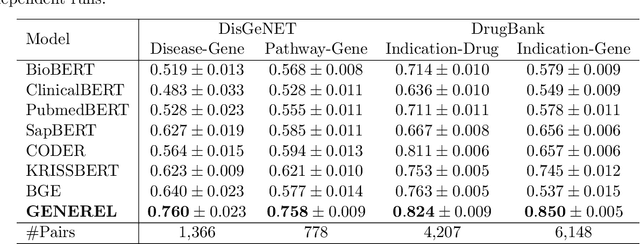
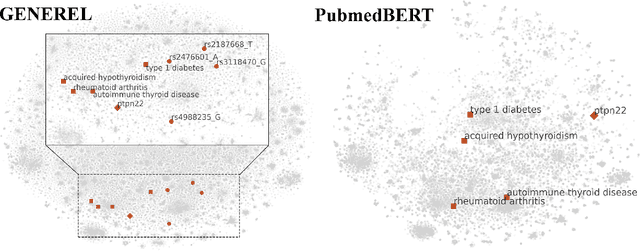
Abstract:We introduce GENomic Encoding REpresentation with Language Model (GENEREL), a framework designed to bridge genetic and biomedical knowledge bases. What sets GENEREL apart is its ability to fine-tune language models to infuse biological knowledge behind clinical concepts such as diseases and medications. This fine-tuning enables the model to capture complex biomedical relationships more effectively, enriching the understanding of how genomic data connects to clinical outcomes. By constructing a unified embedding space for biomedical concepts and a wide range of common SNPs from sources such as patient-level data, biomedical knowledge graphs, and GWAS summaries, GENEREL aligns the embeddings of SNPs and clinical concepts through multi-task contrastive learning. This allows the model to adapt to diverse natural language representations of biomedical concepts while bypassing the limitations of traditional code mapping systems across different data sources. Our experiments demonstrate GENEREL's ability to effectively capture the nuanced relationships between SNPs and clinical concepts. GENEREL also emerges to discern the degree of relatedness, potentially allowing for a more refined identification of concepts. This pioneering approach in constructing a unified embedding system for both SNPs and biomedical concepts enhances the potential for data integration and discovery in biomedical research.
Multi-view Banded Spectral Clustering with Application to ICD9 Clustering
Jun 20, 2018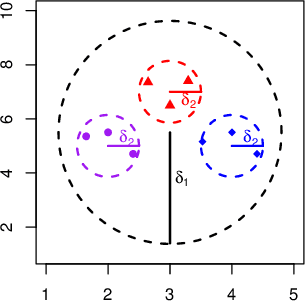
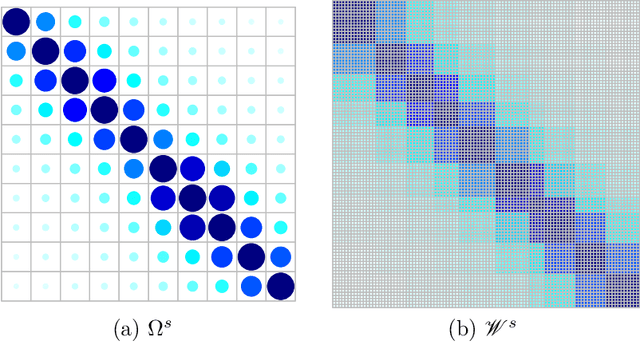
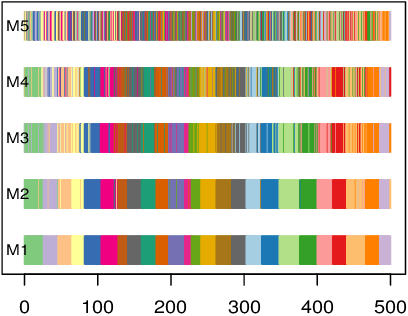
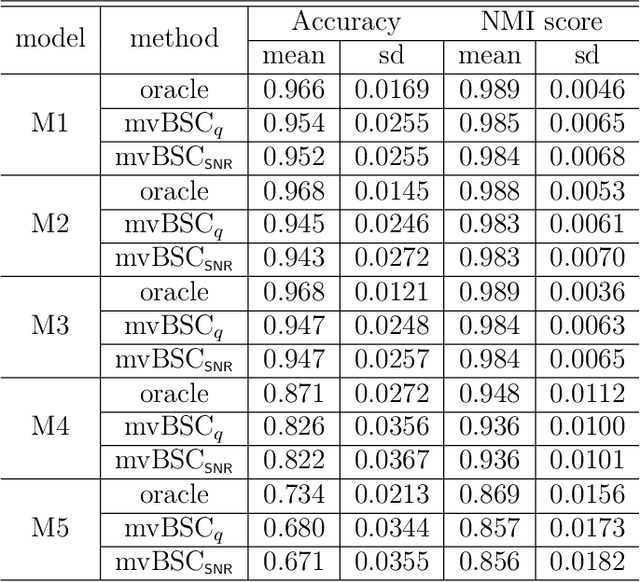
Abstract:Despite recent development in methodology, community detection remains a challenging problem. Existing literature largely focuses on the standard setting where a network is learned using an observed adjacency matrix from a single data source. Constructing a shared network from multiple data sources is more challenging due to the heterogeneity across populations. Additionally, no existing method leverages the prior distance knowledge available in many domains to help the discovery of the network structure. To bridge this gap, in this paper we propose a novel spectral clustering method that optimally combines multiple data sources while leveraging the prior distance knowledge. The proposed method combines a banding step guided by the distance knowledge with a subsequent weighting step to maximize consensus across multiple sources. Its statistical performance is thoroughly studied under a multi-view stochastic block model. We also provide a simple yet optimal rule of choosing weights in practice. The efficacy and robustness of the method is fully demonstrated through extensive simulations. Finally, we apply the method to cluster the International classification of diseases, ninth revision (ICD9), codes and yield a very insightful clustering structure by integrating information from a large claim database and two healthcare systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge