Xujiong Ye
CAPRI-CT: Causal Analysis and Predictive Reasoning for Image Quality Optimization in Computed Tomography
Jul 23, 2025Abstract:In computed tomography (CT), achieving high image quality while minimizing radiation exposure remains a key clinical challenge. This paper presents CAPRI-CT, a novel causal-aware deep learning framework for Causal Analysis and Predictive Reasoning for Image Quality Optimization in CT imaging. CAPRI-CT integrates image data with acquisition metadata (such as tube voltage, tube current, and contrast agent types) to model the underlying causal relationships that influence image quality. An ensemble of Variational Autoencoders (VAEs) is employed to extract meaningful features and generate causal representations from observational data, including CT images and associated imaging parameters. These input features are fused to predict the Signal-to-Noise Ratio (SNR) and support counterfactual inference, enabling what-if simulations, such as changes in contrast agents (types and concentrations) or scan parameters. CAPRI-CT is trained and validated using an ensemble learning approach, achieving strong predictive performance. By facilitating both prediction and interpretability, CAPRI-CT provides actionable insights that could help radiologists and technicians design more efficient CT protocols without repeated physical scans. The source code and dataset are publicly available at https://github.com/SnehaGeorge22/capri-ct.
Deep histological synthesis from mass spectrometry imaging for multimodal registration
Jun 05, 2025
Abstract:Registration of histological and mass spectrometry imaging (MSI) allows for more precise identification of structural changes and chemical interactions in tissue. With histology and MSI having entirely different image formation processes and dimensionalities, registration of the two modalities remains an ongoing challenge. This work proposes a solution that synthesises histological images from MSI, using a pix2pix model, to effectively enable unimodal registration. Preliminary results show promising synthetic histology images with limited artifacts, achieving increases in mutual information (MI) and structural similarity index measures (SSIM) of +0.924 and +0.419, respectively, compared to a baseline U-Net model. Our source code is available on GitHub: https://github.com/kimberley/MIUA2025.
Validation of Human Pose Estimation and Human Mesh Recovery for Extracting Clinically Relevant Motion Data from Videos
Mar 18, 2025Abstract:This work aims to discuss the current landscape of kinematic analysis tools, ranging from the state-of-the-art in sports biomechanics such as inertial measurement units (IMUs) and retroreflective marker-based optical motion capture (MoCap) to more novel approaches from the field of computing such as human pose estimation and human mesh recovery. Primarily, this comparative analysis aims to validate the use of marker-less MoCap techniques in a clinical setting by showing that these marker-less techniques are within a reasonable range for kinematics analysis compared to the more cumbersome and less portable state-of-the-art tools. Not only does marker-less motion capture using human pose estimation produce results in-line with the results of both the IMU and MoCap kinematics but also benefits from a reduced set-up time and reduced practical knowledge and expertise to set up. Overall, while there is still room for improvement when it comes to the quality of the data produced, we believe that this compromise is within the room of error that these low-speed actions that are used in small clinical tests.
MetaFE-DE: Learning Meta Feature Embedding for Depth Estimation from Monocular Endoscopic Images
Feb 05, 2025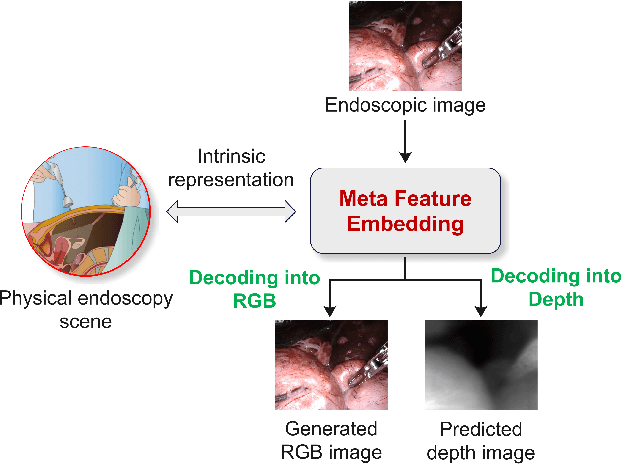
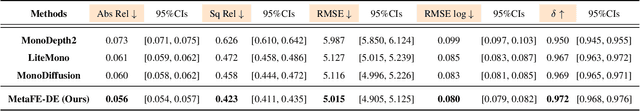
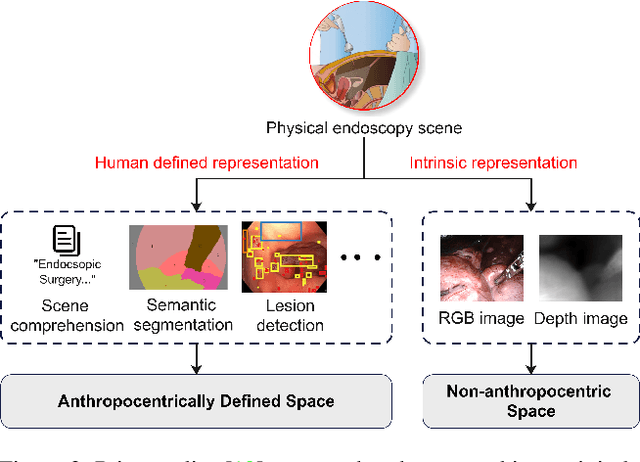
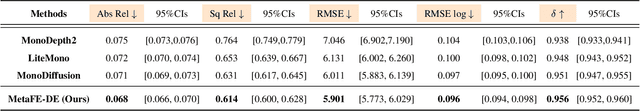
Abstract:Depth estimation from monocular endoscopic images presents significant challenges due to the complexity of endoscopic surgery, such as irregular shapes of human soft tissues, as well as variations in lighting conditions. Existing methods primarily estimate the depth information from RGB images directly, and often surffer the limited interpretability and accuracy. Given that RGB and depth images are two views of the same endoscopic surgery scene, in this paper, we introduce a novel concept referred as ``meta feature embedding (MetaFE)", in which the physical entities (e.g., tissues and surgical instruments) of endoscopic surgery are represented using the shared features that can be alternatively decoded into RGB or depth image. With this concept, we propose a two-stage self-supervised learning paradigm for the monocular endoscopic depth estimation. In the first stage, we propose a temporal representation learner using diffusion models, which are aligned with the spatial information through the cross normalization to construct the MetaFE. In the second stage, self-supervised monocular depth estimation with the brightness calibration is applied to decode the meta features into the depth image. Extensive evaluation on diverse endoscopic datasets demonstrates that our approach outperforms the state-of-the-art method in depth estimation, achieving superior accuracy and generalization. The source code will be publicly available.
Pushing the limits of cell segmentation models for imaging mass cytometry
Feb 06, 2024



Abstract:Imaging mass cytometry (IMC) is a relatively new technique for imaging biological tissue at subcellular resolution. In recent years, learning-based segmentation methods have enabled precise quantification of cell type and morphology, but typically rely on large datasets with fully annotated ground truth (GT) labels. This paper explores the effects of imperfect labels on learning-based segmentation models and evaluates the generalisability of these models to different tissue types. Our results show that removing 50% of cell annotations from GT masks only reduces the dice similarity coefficient (DSC) score to 0.874 (from 0.889 achieved by a model trained on fully annotated GT masks). This implies that annotation time can in fact be reduced by at least half without detrimentally affecting performance. Furthermore, training our single-tissue model on imperfect labels only decreases DSC by 0.031 on an unseen tissue type compared to its multi-tissue counterpart, with negligible qualitative differences in segmentation. Additionally, bootstrapping the worst-performing model (with 5% of cell annotations) a total of ten times improves its original DSC score of 0.720 to 0.829. These findings imply that less time and work can be put into the process of producing comparable segmentation models; this includes eliminating the need for multiple IMC tissue types during training, whilst also providing the potential for models with very few labels to improve on themselves. Source code is available on GitHub: https://github.com/kimberley/ISBI2024.
Rethinking the transfer learning for FCN based polyp segmentation in colonoscopy
Nov 04, 2022Abstract:Besides the complex nature of colonoscopy frames with intrinsic frame formation artefacts such as light reflections and the diversity of polyp types/shapes, the publicly available polyp segmentation training datasets are limited, small and imbalanced. In this case, the automated polyp segmentation using a deep neural network remains an open challenge due to the overfitting of training on small datasets. We proposed a simple yet effective polyp segmentation pipeline that couples the segmentation (FCN) and classification (CNN) tasks. We find the effectiveness of interactive weight transfer between dense and coarse vision tasks that mitigates the overfitting in learning. And It motivates us to design a new training scheme within our segmentation pipeline. Our method is evaluated on CVC-EndoSceneStill and Kvasir-SEG datasets. It achieves 4.34% and 5.70% Polyp-IoU improvements compared to the state-of-the-art methods on the EndoSceneStill and Kvasir-SEG datasets, respectively.
Dual-stream spatiotemporal networks with feature sharing for monitoring animals in the home cage
Jun 01, 2022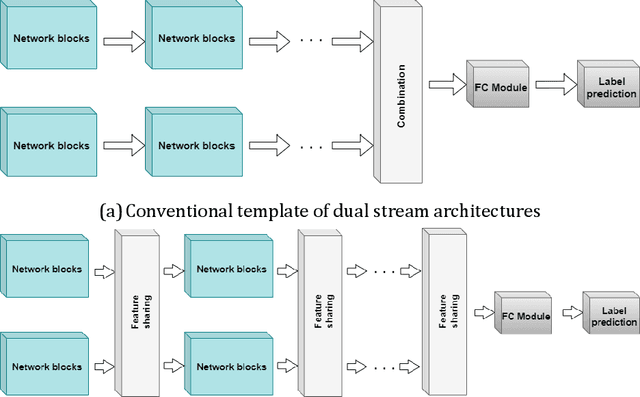
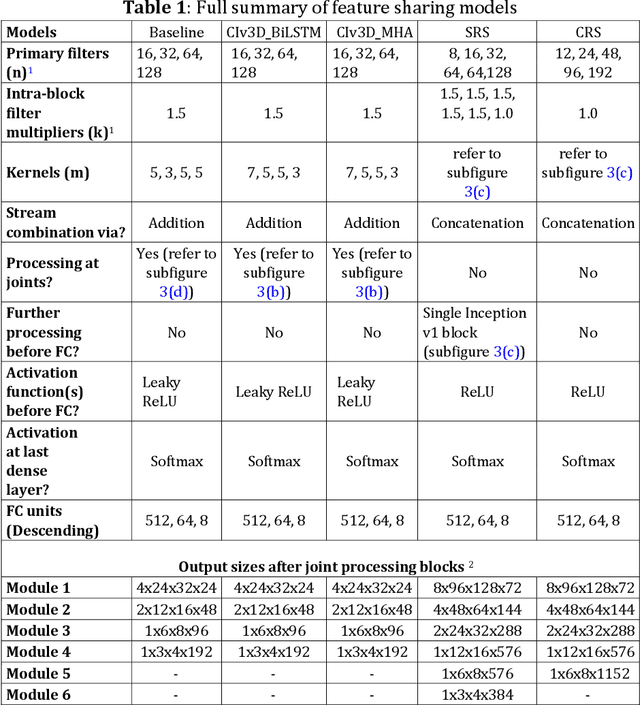
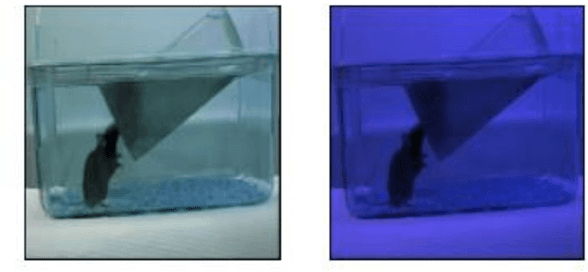
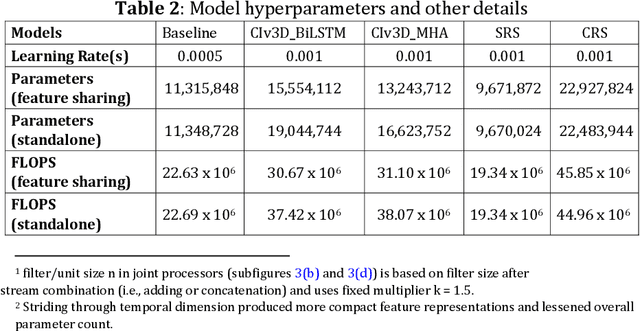
Abstract:This paper presents a spatiotemporal deep learning approach for mouse behavioural classification in the home cage. Using a series of dual-stream architectures with assorted modifications to increase performance, we introduce a novel feature-sharing approach that jointly processes the streams at regular intervals throughout the network. Using a publicly available labelled dataset of singly-housed mice, we achieve a prediction accuracy of 86.47% using an ensemble of Inception-based networks that utilize feature sharing. We also demonstrate through ablation studies that for all models, the feature-sharing architectures consistently perform better than conventional ones having separate streams. The best performing models were further evaluated on other activity datasets, both mouse and human, and achieved state-of-the-art results. Future work will investigate the effectiveness of feature sharing in behavioural classification in the unsupervised anomaly detection domain.
Unsupervised detection of mouse behavioural anomalies using two-stream convolutional autoencoders
May 28, 2021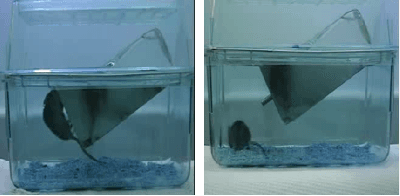
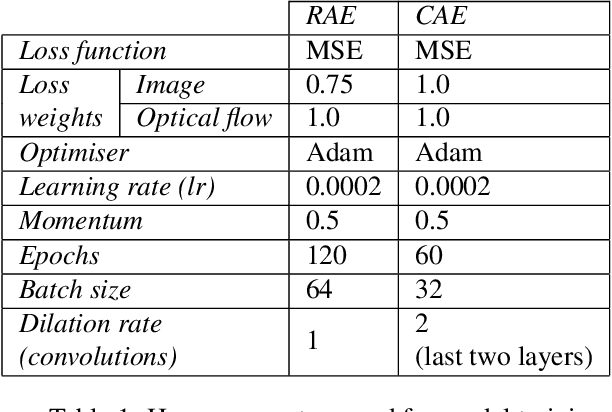
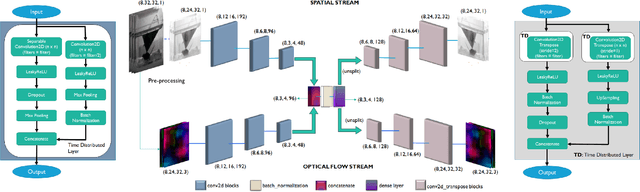
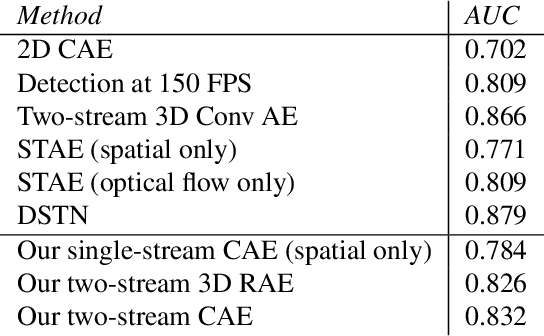
Abstract:This paper explores the application of unsupervised learning to detecting anomalies in mouse video data. The two models presented in this paper are a dual-stream, 3D convolutional autoencoder (with residual connections) and a dual-stream, 2D convolutional autoencoder. The publicly available dataset used here contains twelve videos of single home-caged mice alongside frame-level annotations. Under the pretext that the autoencoder only sees normal events, the video data was handcrafted to treat each behaviour as a pseudo-anomaly thereby eliminating them from the others during training. The results are presented for one conspicuous behaviour (hang) and one inconspicuous behaviour (groom). The performance of these models is compared to a single stream autoencoder and a supervised learning model, which are both based on the custom CAE. Both models are also tested on the CUHK Avenue dataset were found to perform as well as some state-of-the-art architectures.
DR-Unet104 for Multimodal MRI brain tumor segmentation
Nov 04, 2020



Abstract:In this paper we propose a 2D deep residual Unet with 104 convolutional layers (DR-Unet104) for lesion segmentation in brain MRIs. We make multiple additions to the Unet architecture, including adding the 'bottleneck' residual block to the Unet encoder and adding dropout after each convolution block stack. We verified the effect of introducing the regularisation of dropout with small rate (e.g. 0.2) on the architecture, and found a dropout of 0.2 improved the overall performance compared to no dropout, or a dropout of 0.5. We evaluated the proposed architecture as part of the Multimodal Brain Tumor Segmentation (BraTS) 2020 Challenge and compared our method to DeepLabV3+ with a ResNet-V2-152 backbone. We found that the DR-Unet104 achieved a mean dice score coefficient of 0.8862, 0.6756 and 0.6721 for validation data, whole tumor, enhancing tumor and tumor core respectively, an overall improvement on 0.8770, 0.65242 and 0.68134 achieved by DeepLabV3+. Our method produced a final mean DSC of 0.8673, 0.7514 and 0.7983 on whole tumor, enhancing tumor and tumor core on the challenge's testing data. We present this as a state-of-the-art 2D lesion segmentation architecture that can be used on lower power computers than a 3D architecture. The source code and trained model for this work is openly available at https://github.com/jordan-colman/DR-Unet104.
MRI Brain Tumor Segmentation using Random Forests and Fully Convolutional Networks
Sep 13, 2019


Abstract:In this paper, we propose a novel learning based method for automated segmentation of brain tumor in multimodal MRI images, which incorporates two sets of machine -learned and hand crafted features. Fully convolutional networks (FCN) forms the machine learned features and texton based features are considered as hand-crafted features. Random forest (RF) is used to classify the MRI image voxels into normal brain tissues and different parts of tumors, i.e. edema, necrosis and enhancing tumor. The method was evaluated on BRATS 2017 challenge dataset. The results show that the proposed method provides promising segmentations. The mean Dice overlap measure for automatic brain tumor segmentation against ground truth is 0.86, 0.78 and 0.66 for whole tumor, core and enhancing tumor, respectively.
* Published in the pre-conference proceeding of "2017 International MICCAI BraTS Challenge"
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge