Luc Bidaut
Dual-stream spatiotemporal networks with feature sharing for monitoring animals in the home cage
Jun 01, 2022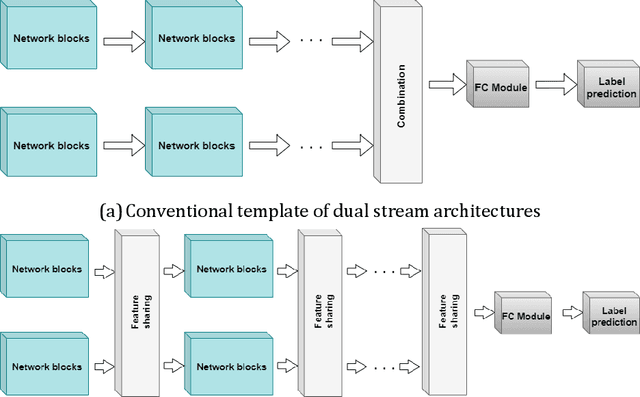
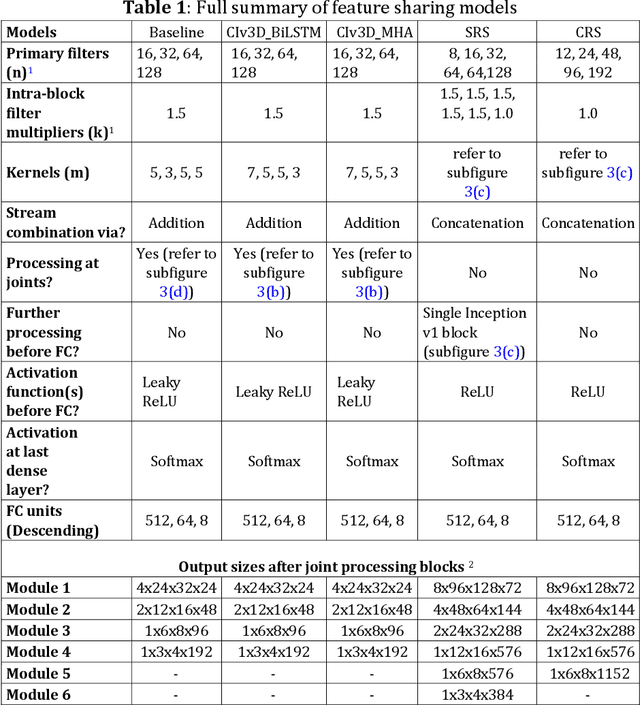
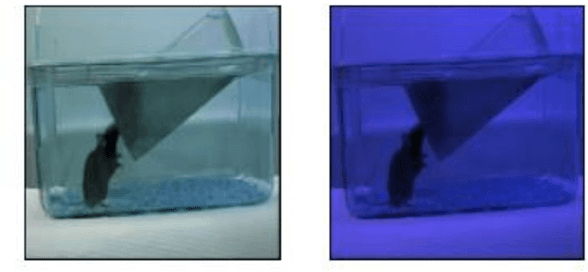
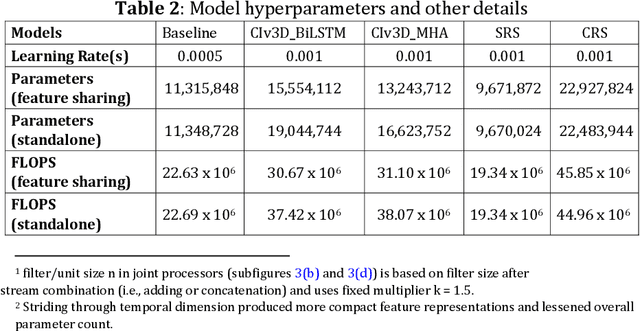
Abstract:This paper presents a spatiotemporal deep learning approach for mouse behavioural classification in the home cage. Using a series of dual-stream architectures with assorted modifications to increase performance, we introduce a novel feature-sharing approach that jointly processes the streams at regular intervals throughout the network. Using a publicly available labelled dataset of singly-housed mice, we achieve a prediction accuracy of 86.47% using an ensemble of Inception-based networks that utilize feature sharing. We also demonstrate through ablation studies that for all models, the feature-sharing architectures consistently perform better than conventional ones having separate streams. The best performing models were further evaluated on other activity datasets, both mouse and human, and achieved state-of-the-art results. Future work will investigate the effectiveness of feature sharing in behavioural classification in the unsupervised anomaly detection domain.
Unsupervised detection of mouse behavioural anomalies using two-stream convolutional autoencoders
May 28, 2021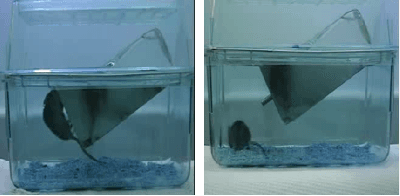
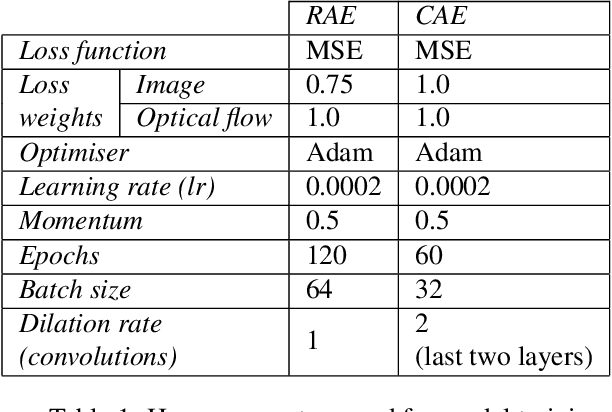
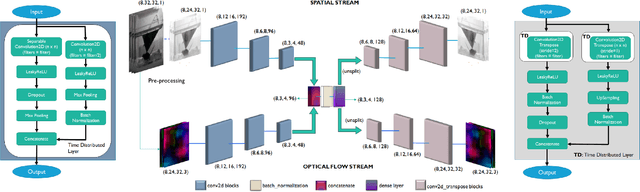
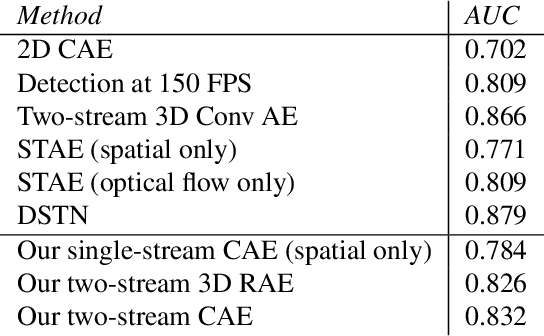
Abstract:This paper explores the application of unsupervised learning to detecting anomalies in mouse video data. The two models presented in this paper are a dual-stream, 3D convolutional autoencoder (with residual connections) and a dual-stream, 2D convolutional autoencoder. The publicly available dataset used here contains twelve videos of single home-caged mice alongside frame-level annotations. Under the pretext that the autoencoder only sees normal events, the video data was handcrafted to treat each behaviour as a pseudo-anomaly thereby eliminating them from the others during training. The results are presented for one conspicuous behaviour (hang) and one inconspicuous behaviour (groom). The performance of these models is compared to a single stream autoencoder and a supervised learning model, which are both based on the custom CAE. Both models are also tested on the CUHK Avenue dataset were found to perform as well as some state-of-the-art architectures.
A Unified Deep Learning Approach for Prediction of Parkinson's Disease
Nov 25, 2019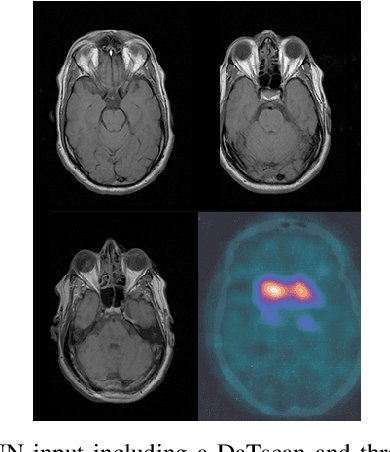

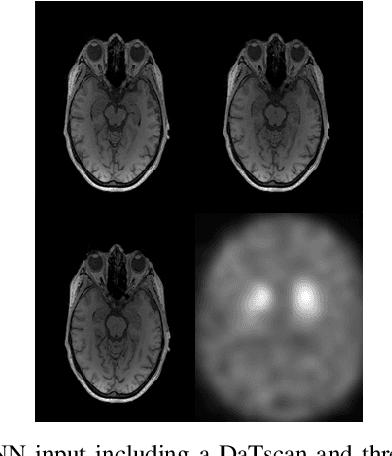
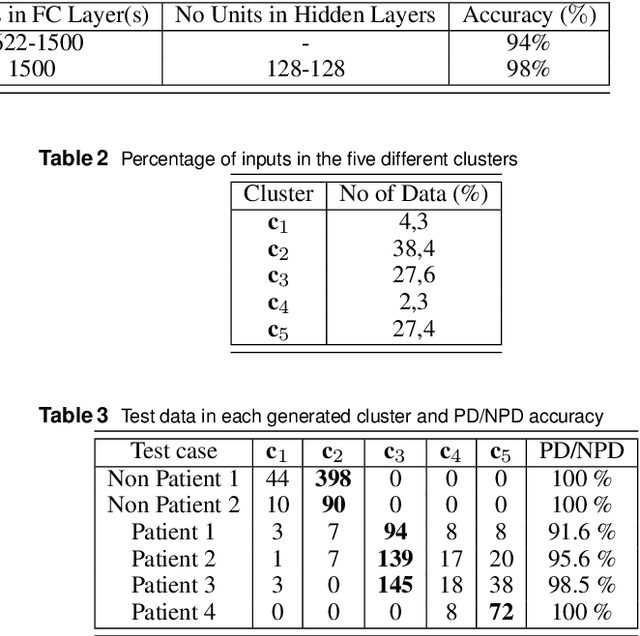
Abstract:The paper presents a novel approach, based on deep learning, for diagnosis of Parkinson's disease through medical imaging. The approach includes analysis and use of the knowledge extracted by Deep Convolutional and Recurrent Neural Networks (DNNs) when trained with medical images, such as Magnetic Resonance Images and DaTscans. Internal representations of the trained DNNs constitute the extracted knowledge which is used in a transfer learning and domain adaptation manner, so as to create a unified framework for prediction of Parkinson's across different medical environments. A large experimental study is presented illustrating the ability of the proposed approach to effectively predict Parkinson's, using different medical image sets from real environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge