Yue Leng
Sky-Drive: A Distributed Multi-Agent Simulation Platform for Socially-Aware and Human-AI Collaborative Future Transportation
Apr 25, 2025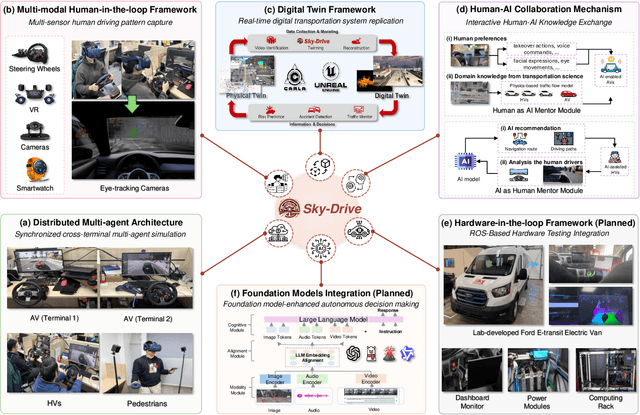
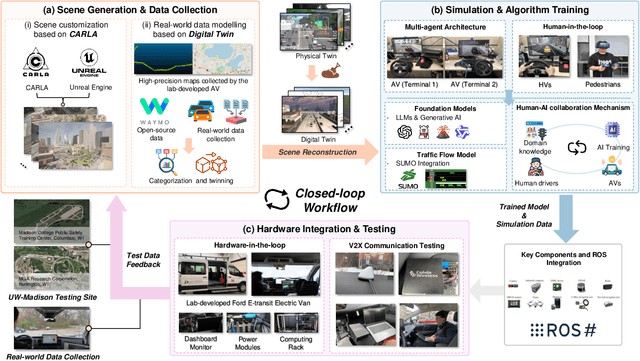
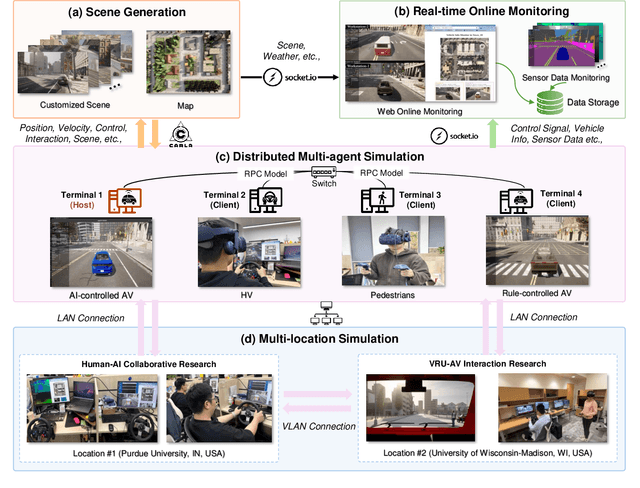
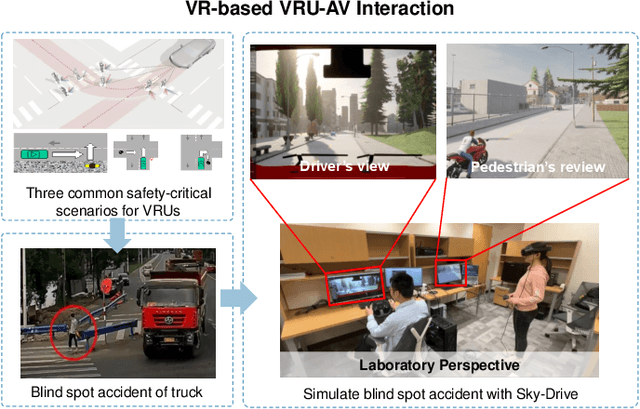
Abstract:Recent advances in autonomous system simulation platforms have significantly enhanced the safe and scalable testing of driving policies. However, existing simulators do not yet fully meet the needs of future transportation research, particularly in modeling socially-aware driving agents and enabling effective human-AI collaboration. This paper introduces Sky-Drive, a novel distributed multi-agent simulation platform that addresses these limitations through four key innovations: (a) a distributed architecture for synchronized simulation across multiple terminals; (b) a multi-modal human-in-the-loop framework integrating diverse sensors to collect rich behavioral data; (c) a human-AI collaboration mechanism supporting continuous and adaptive knowledge exchange; and (d) a digital twin (DT) framework for constructing high-fidelity virtual replicas of real-world transportation environments. Sky-Drive supports diverse applications such as autonomous vehicle (AV)-vulnerable road user (VRU) interaction modeling, human-in-the-loop training, socially-aware reinforcement learning, personalized driving policy, and customized scenario generation. Future extensions will incorporate foundation models for context-aware decision support and hardware-in-the-loop (HIL) testing for real-world validation. By bridging scenario generation, data collection, algorithm training, and hardware integration, Sky-Drive has the potential to become a foundational platform for the next generation of socially-aware and human-centered autonomous transportation research. The demo video and code are available at:https://sky-lab-uw.github.io/Sky-Drive-website/
CurricuVLM: Towards Safe Autonomous Driving via Personalized Safety-Critical Curriculum Learning with Vision-Language Models
Feb 21, 2025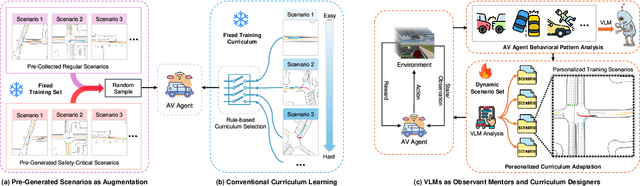
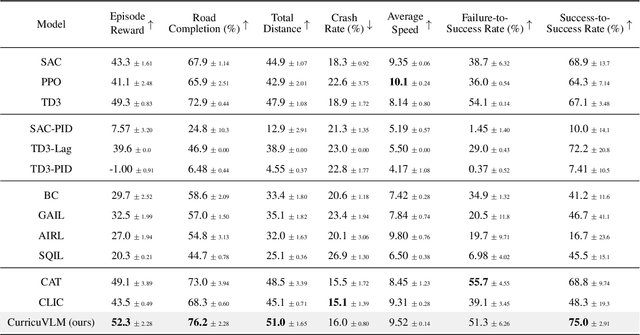
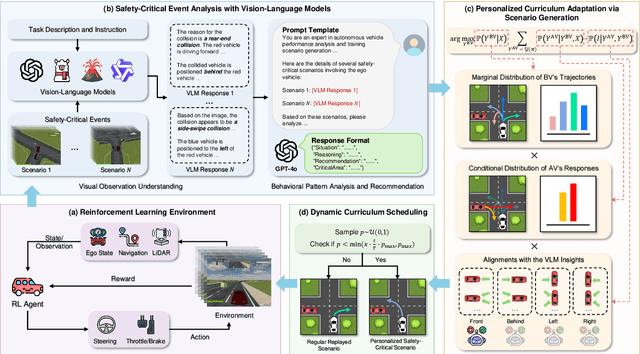
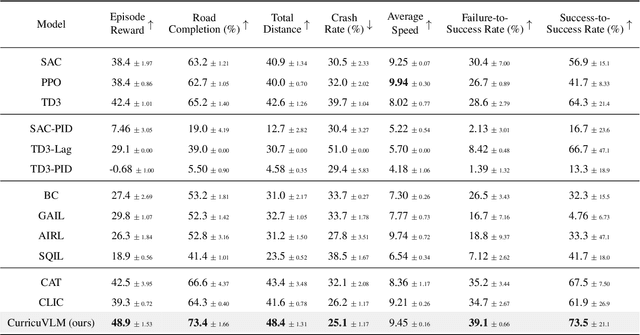
Abstract:Ensuring safety in autonomous driving systems remains a critical challenge, particularly in handling rare but potentially catastrophic safety-critical scenarios. While existing research has explored generating safety-critical scenarios for autonomous vehicle (AV) testing, there is limited work on effectively incorporating these scenarios into policy learning to enhance safety. Furthermore, developing training curricula that adapt to an AV's evolving behavioral patterns and performance bottlenecks remains largely unexplored. To address these challenges, we propose CurricuVLM, a novel framework that leverages Vision-Language Models (VLMs) to enable personalized curriculum learning for autonomous driving agents. Our approach uniquely exploits VLMs' multimodal understanding capabilities to analyze agent behavior, identify performance weaknesses, and dynamically generate tailored training scenarios for curriculum adaptation. Through comprehensive analysis of unsafe driving situations with narrative descriptions, CurricuVLM performs in-depth reasoning to evaluate the AV's capabilities and identify critical behavioral patterns. The framework then synthesizes customized training scenarios targeting these identified limitations, enabling effective and personalized curriculum learning. Extensive experiments on the Waymo Open Motion Dataset show that CurricuVLM outperforms state-of-the-art baselines across both regular and safety-critical scenarios, achieving superior performance in terms of navigation success, driving efficiency, and safety metrics. Further analysis reveals that CurricuVLM serves as a general approach that can be integrated with various RL algorithms to enhance autonomous driving systems. The code and demo video are available at: https://zihaosheng.github.io/CurricuVLM/.
SleepNetZero: Zero-Burden Zero-Shot Reliable Sleep Staging With Neural Networks Based on Ballistocardiograms
Oct 30, 2024Abstract:Sleep monitoring plays a crucial role in maintaining good health, with sleep staging serving as an essential metric in the monitoring process. Traditional methods, utilizing medical sensors like EEG and ECG, can be effective but often present challenges such as unnatural user experience, complex deployment, and high costs. Ballistocardiography~(BCG), a type of piezoelectric sensor signal, offers a non-invasive, user-friendly, and easily deployable alternative for long-term home monitoring. However, reliable BCG-based sleep staging is challenging due to the limited sleep monitoring data available for BCG. A restricted training dataset prevents the model from generalization across populations. Additionally, transferring to BCG faces difficulty ensuring model robustness when migrating from other data sources. To address these issues, we introduce SleepNetZero, a zero-shot learning based approach for sleep staging. To tackle the generalization challenge, we propose a series of BCG feature extraction methods that align BCG components with corresponding respiratory, cardiac, and movement channels in PSG. This allows models to be trained on large-scale PSG datasets that are diverse in population. For the migration challenge, we employ data augmentation techniques, significantly enhancing generalizability. We conducted extensive training and testing on large datasets~(12393 records from 9637 different subjects), achieving an accuracy of 0.803 and a Cohen's Kappa of 0.718. ZeroSleepNet was also deployed in real prototype~(monitoring pads) and tested in actual hospital settings~(265 users), demonstrating an accuracy of 0.697 and a Cohen's Kappa of 0.589. To the best of our knowledge, this work represents the first known reliable BCG-based sleep staging effort and marks a significant step towards in-home health monitoring.
Annotation of Sleep Depth Index with Scalable Deep Learning Yields Novel Digital Biomarkers for Sleep Health
Jul 05, 2024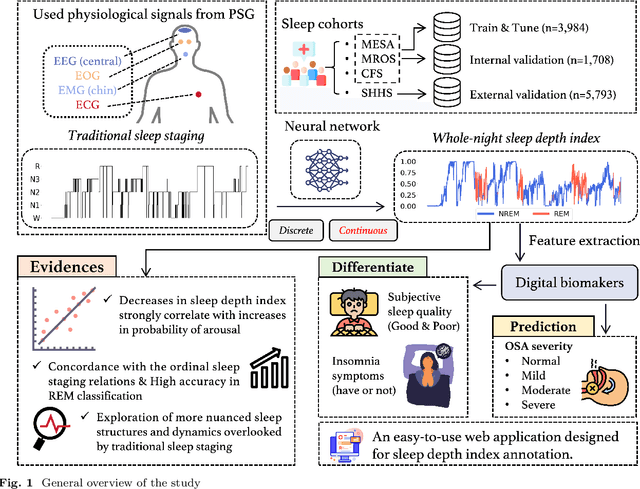

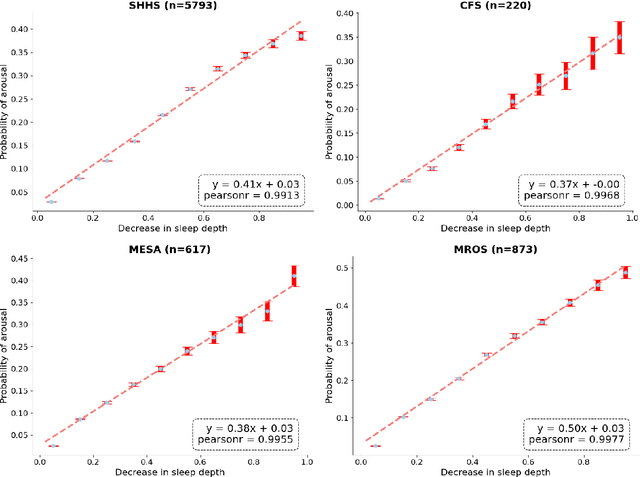
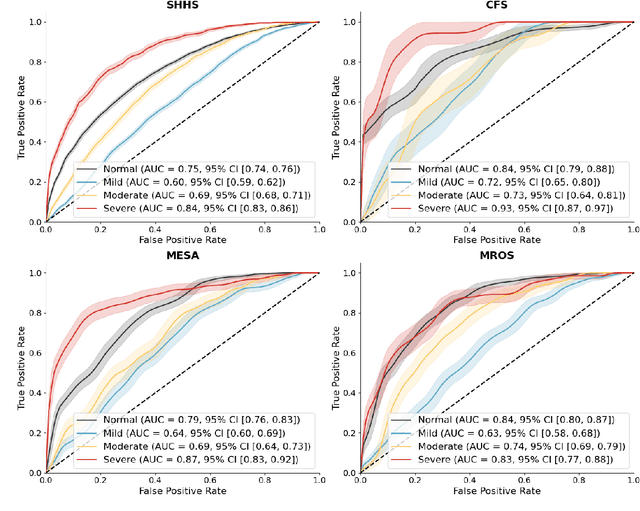
Abstract:Traditional sleep staging categorizes sleep and wakefulness into five coarse-grained classes, overlooking subtle variations within each stage. It provides limited information about the probability of arousal and may hinder the diagnosis of sleep disorders, such as insomnia. To address this issue, we propose a deep-learning method for automatic and scalable annotation of sleep depth index using existing sleep staging labels. Our approach is validated using polysomnography from over ten thousand recordings across four large-scale cohorts. The results show a strong correlation between the decrease in sleep depth index and the increase in arousal likelihood. Several case studies indicate that the sleep depth index captures more nuanced sleep structures than conventional sleep staging. Sleep biomarkers extracted from the whole-night sleep depth index exhibit statistically significant differences with medium-to-large effect sizes across groups of varied subjective sleep quality and insomnia symptoms. These sleep biomarkers also promise utility in predicting the severity of obstructive sleep apnea, particularly in severe cases. Our study underscores the utility of the proposed method for continuous sleep depth annotation, which could reveal more detailed structures and dynamics within whole-night sleep and yield novel digital biomarkers beneficial for sleep health.
Predicting the Mechanical Properties of Fibrin Using Neural Networks Trained on Discrete Fiber Network Data
Jan 23, 2021



Abstract:Fibrin is a structural protein key for processes such as wound healing and thrombus formation. At the macroscale, fibrin forms a gel and has a mechanical response that is dictated by the mechanics of a microscale fiber network. Hence, accurate description of fibrin gels can be achieved using representative volume elements (RVE) that explicitly model the discrete fiber networks of the microscale. These RVE models, however, cannot be efficiently used to model the macroscale due to the challenges and computational demands of multiscale coupling. Here, we propose the use of an artificial, fully connected neural network (FCNN) to efficiently capture the behavior of the RVE models. The FCNN was trained on 1100 fiber networks subjected to 121 biaxial deformations. The stress data from the RVE, together with the total energy on the fibers and the condition of incompressibility of the surrounding matrix, were used to determine the derivatives of an unknown strain energy function with respect to the deformation invariants. During training, the loss function was modified to ensure convexity of the strain energy function and symmetry of its Hessian. A general FCNN model was coded into a user material subroutine (UMAT) in the software Abaqus. The UMAT implementation takes in the structure and parameters of an arbitrary FCNN as material parameters from the input file. The inputs to the FCNN include the first two isochoric invariants of the deformation. The FCNN outputs the derivatives of the strain energy with respect to the isochoric invariants. In this work, the FCNN trained on the discrete fiber network data was used in finite element simulations of fibrin gels using our UMAT. We anticipate that this work will enable further integration of machine learning tools with computational mechanics. It will also improve computational modeling of biological materials characterized by a multiscale structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge