Weijun Huang
SleepNetZero: Zero-Burden Zero-Shot Reliable Sleep Staging With Neural Networks Based on Ballistocardiograms
Oct 30, 2024Abstract:Sleep monitoring plays a crucial role in maintaining good health, with sleep staging serving as an essential metric in the monitoring process. Traditional methods, utilizing medical sensors like EEG and ECG, can be effective but often present challenges such as unnatural user experience, complex deployment, and high costs. Ballistocardiography~(BCG), a type of piezoelectric sensor signal, offers a non-invasive, user-friendly, and easily deployable alternative for long-term home monitoring. However, reliable BCG-based sleep staging is challenging due to the limited sleep monitoring data available for BCG. A restricted training dataset prevents the model from generalization across populations. Additionally, transferring to BCG faces difficulty ensuring model robustness when migrating from other data sources. To address these issues, we introduce SleepNetZero, a zero-shot learning based approach for sleep staging. To tackle the generalization challenge, we propose a series of BCG feature extraction methods that align BCG components with corresponding respiratory, cardiac, and movement channels in PSG. This allows models to be trained on large-scale PSG datasets that are diverse in population. For the migration challenge, we employ data augmentation techniques, significantly enhancing generalizability. We conducted extensive training and testing on large datasets~(12393 records from 9637 different subjects), achieving an accuracy of 0.803 and a Cohen's Kappa of 0.718. ZeroSleepNet was also deployed in real prototype~(monitoring pads) and tested in actual hospital settings~(265 users), demonstrating an accuracy of 0.697 and a Cohen's Kappa of 0.589. To the best of our knowledge, this work represents the first known reliable BCG-based sleep staging effort and marks a significant step towards in-home health monitoring.
FetusMapV2: Enhanced Fetal Pose Estimation in 3D Ultrasound
Oct 30, 2023Abstract:Fetal pose estimation in 3D ultrasound (US) involves identifying a set of associated fetal anatomical landmarks. Its primary objective is to provide comprehensive information about the fetus through landmark connections, thus benefiting various critical applications, such as biometric measurements, plane localization, and fetal movement monitoring. However, accurately estimating the 3D fetal pose in US volume has several challenges, including poor image quality, limited GPU memory for tackling high dimensional data, symmetrical or ambiguous anatomical structures, and considerable variations in fetal poses. In this study, we propose a novel 3D fetal pose estimation framework (called FetusMapV2) to overcome the above challenges. Our contribution is three-fold. First, we propose a heuristic scheme that explores the complementary network structure-unconstrained and activation-unreserved GPU memory management approaches, which can enlarge the input image resolution for better results under limited GPU memory. Second, we design a novel Pair Loss to mitigate confusion caused by symmetrical and similar anatomical structures. It separates the hidden classification task from the landmark localization task and thus progressively eases model learning. Last, we propose a shape priors-based self-supervised learning by selecting the relatively stable landmarks to refine the pose online. Extensive experiments and diverse applications on a large-scale fetal US dataset including 1000 volumes with 22 landmarks per volume demonstrate that our method outperforms other strong competitors.
Agent with Tangent-based Formulation and Anatomical Perception for Standard Plane Localization in 3D Ultrasound
Jul 01, 2022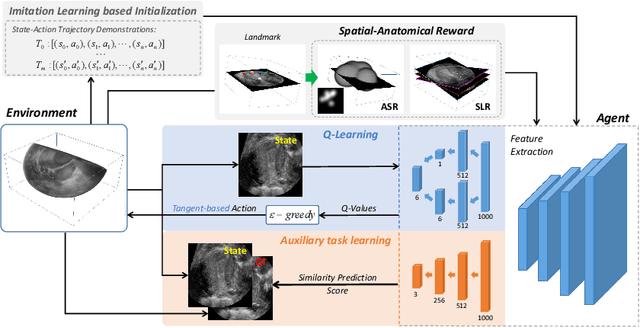
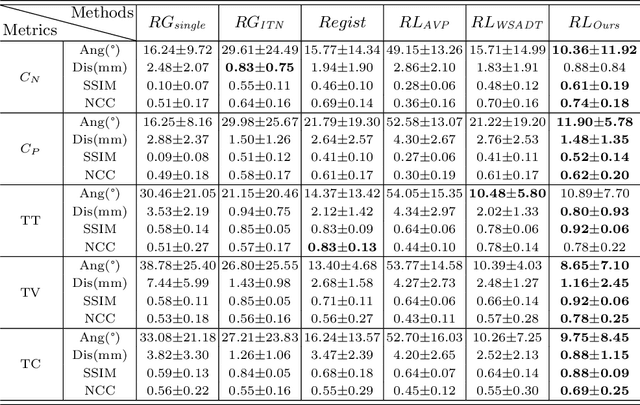
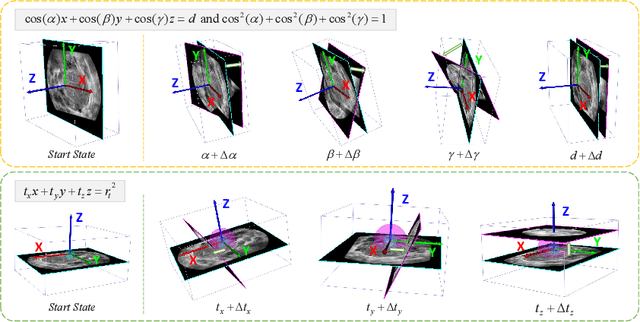
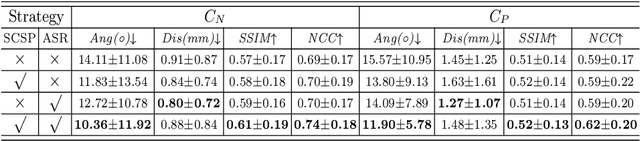
Abstract:Standard plane (SP) localization is essential in routine clinical ultrasound (US) diagnosis. Compared to 2D US, 3D US can acquire multiple view planes in one scan and provide complete anatomy with the addition of coronal plane. However, manually navigating SPs in 3D US is laborious and biased due to the orientation variability and huge search space. In this study, we introduce a novel reinforcement learning (RL) framework for automatic SP localization in 3D US. Our contribution is three-fold. First, we formulate SP localization in 3D US as a tangent-point-based problem in RL to restructure the action space and significantly reduce the search space. Second, we design an auxiliary task learning strategy to enhance the model's ability to recognize subtle differences crossing Non-SPs and SPs in plane search. Finally, we propose a spatial-anatomical reward to effectively guide learning trajectories by exploiting spatial and anatomical information simultaneously. We explore the efficacy of our approach on localizing four SPs on uterus and fetal brain datasets. The experiments indicate that our approach achieves a high localization accuracy as well as robust performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge