Alina Jade Barnett
It's LIT! Reliability-Optimized LLMs with Inspectable Tools
Nov 18, 2025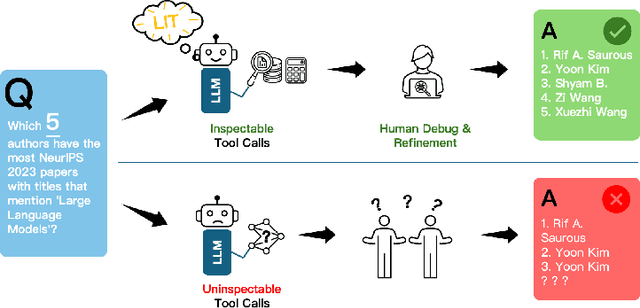
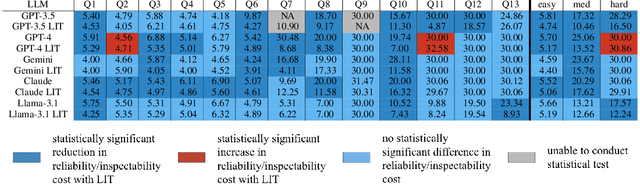
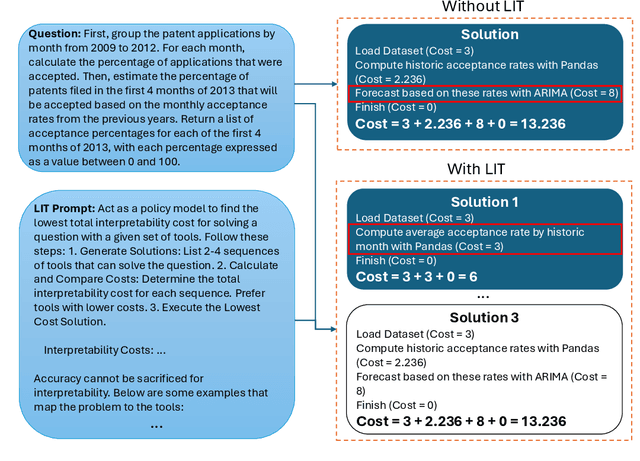
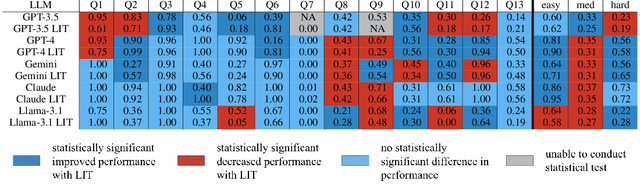
Abstract:Large language models (LLMs) have exhibited remarkable capabilities across various domains. The ability to call external tools further expands their capability to handle real-world tasks. However, LLMs often follow an opaque reasoning process, which limits their usefulness in high-stakes domains where solutions need to be trustworthy to end users. LLMs can choose solutions that are unreliable and difficult to troubleshoot, even if better options are available. We address this issue by forcing LLMs to use external -- more reliable -- tools to solve problems when possible. We present a framework built on the tool-calling capabilities of existing LLMs to enable them to select the most reliable and easy-to-troubleshoot solution path, which may involve multiple sequential tool calls. We refer to this framework as LIT (LLMs with Inspectable Tools). In order to support LIT, we introduce a new and challenging benchmark dataset of 1,300 questions and a customizable set of reliability cost functions associated with a collection of specialized tools. These cost functions summarize how reliable each tool is and how easy it is to troubleshoot. For instance, a calculator is reliable across domains, whereas a linear prediction model is not reliable if there is distribution shift, but it is easy to troubleshoot. A tool that constructs a random forest is neither reliable nor easy to troubleshoot. These tools interact with the Harvard USPTO Patent Dataset and a new dataset of NeurIPS 2023 papers to solve mathematical, coding, and modeling problems of varying difficulty levels. We demonstrate that LLMs can achieve more reliable and informed problem-solving while maintaining task performance using our framework.
Rashomon Sets for Prototypical-Part Networks: Editing Interpretable Models in Real-Time
Mar 03, 2025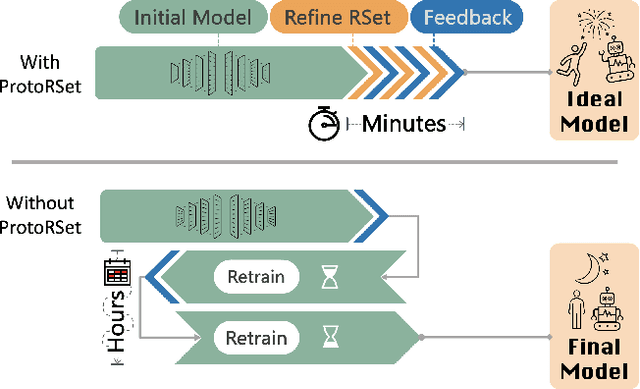
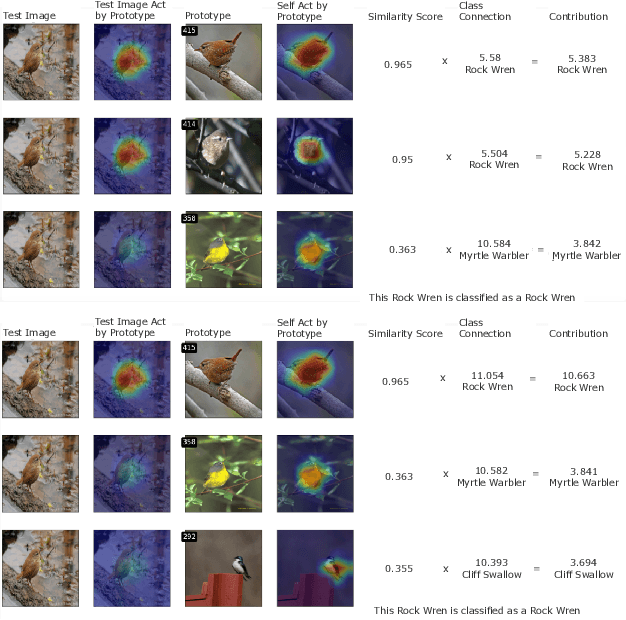
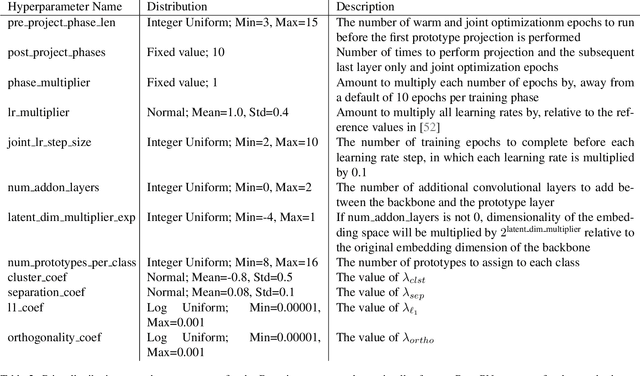
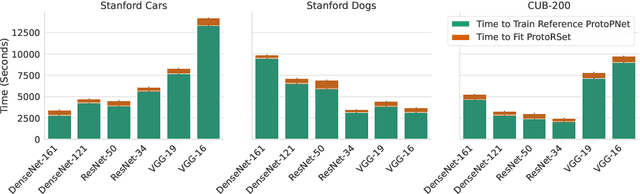
Abstract:Interpretability is critical for machine learning models in high-stakes settings because it allows users to verify the model's reasoning. In computer vision, prototypical part models (ProtoPNets) have become the dominant model type to meet this need. Users can easily identify flaws in ProtoPNets, but fixing problems in a ProtoPNet requires slow, difficult retraining that is not guaranteed to resolve the issue. This problem is called the "interaction bottleneck." We solve the interaction bottleneck for ProtoPNets by simultaneously finding many equally good ProtoPNets (i.e., a draw from a "Rashomon set"). We show that our framework - called Proto-RSet - quickly produces many accurate, diverse ProtoPNets, allowing users to correct problems in real time while maintaining performance guarantees with respect to the training set. We demonstrate the utility of this method in two settings: 1) removing synthetic bias introduced to a bird identification model and 2) debugging a skin cancer identification model. This tool empowers non-machine-learning experts, such as clinicians or domain experts, to quickly refine and correct machine learning models without repeated retraining by machine learning experts.
This Looks Better than That: Better Interpretable Models with ProtoPNeXt
Jun 20, 2024



Abstract:Prototypical-part models are a popular interpretable alternative to black-box deep learning models for computer vision. However, they are difficult to train, with high sensitivity to hyperparameter tuning, inhibiting their application to new datasets and our understanding of which methods truly improve their performance. To facilitate the careful study of prototypical-part networks (ProtoPNets), we create a new framework for integrating components of prototypical-part models -- ProtoPNeXt. Using ProtoPNeXt, we show that applying Bayesian hyperparameter tuning and an angular prototype similarity metric to the original ProtoPNet is sufficient to produce new state-of-the-art accuracy for prototypical-part models on CUB-200 across multiple backbones. We further deploy this framework to jointly optimize for accuracy and prototype interpretability as measured by metrics included in ProtoPNeXt. Using the same resources, this produces models with substantially superior semantics and changes in accuracy between +1.3% and -1.5%. The code and trained models will be made publicly available upon publication.
FPN-IAIA-BL: A Multi-Scale Interpretable Deep Learning Model for Classification of Mass Margins in Digital Mammography
Jun 10, 2024



Abstract:Digital mammography is essential to breast cancer detection, and deep learning offers promising tools for faster and more accurate mammogram analysis. In radiology and other high-stakes environments, uninterpretable ("black box") deep learning models are unsuitable and there is a call in these fields to make interpretable models. Recent work in interpretable computer vision provides transparency to these formerly black boxes by utilizing prototypes for case-based explanations, achieving high accuracy in applications including mammography. However, these models struggle with precise feature localization, reasoning on large portions of an image when only a small part is relevant. This paper addresses this gap by proposing a novel multi-scale interpretable deep learning model for mammographic mass margin classification. Our contribution not only offers an interpretable model with reasoning aligned with radiologist practices, but also provides a general architecture for computer vision with user-configurable prototypes from coarse- to fine-grained prototypes.
Mapping the Ictal-Interictal-Injury Continuum Using Interpretable Machine Learning
Nov 14, 2022


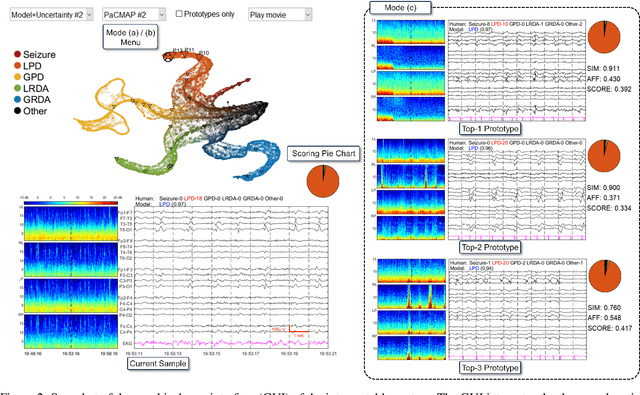
Abstract:IMPORTANCE: An interpretable machine learning model can provide faithful explanations of each prediction and yet maintain higher performance than its black box counterpart. OBJECTIVE: To design an interpretable machine learning model which accurately predicts EEG protopatterns while providing an explanation of its predictions with assistance of a specialized GUI. To map the cEEG latent features to a 2D space in order to visualize the ictal-interictal-injury continuum and gain insight into its high-dimensional structure. DESIGN, SETTING, AND PARTICIPANTS: 50,697 50-second cEEG samples from 2,711 ICU patients collected between July 2006 and March 2020 at Massachusetts General Hospital. Samples were labeled as one of 6 EEG activities by domain experts, with 124 different experts providing annotations. MAIN OUTCOMES AND MEASURES: Our neural network is interpretable because it uses case-based reasoning: it compares a new EEG reading to a set of learned prototypical EEG samples from the training dataset. Interpretability was measured with task-specific neighborhood agreement statistics. Discriminatory performance was evaluated with AUROC and AUPRC. RESULTS: The model achieves AUROCs of 0.87, 0.93, 0.96, 0.92, 0.93, 0.80 for classes Seizure, LPD, GPD, LRDA, GRDA, Other respectively. This performance is statistically significantly higher than that of the corresponding uninterpretable (black box) model with p<0.0001. Videos of the ictal-interictal-injury continuum are provided. CONCLUSION AND RELEVANCE: Our interpretable model and GUI can act as a reference for practitioners who work with cEEG patterns. We can now better understand the relationships between different types of cEEG patterns. In the future, this system may allow for targeted intervention and training in clinical settings. It could also be used for re-confirming or providing additional information for diagnostics.
Deformable ProtoPNet: An Interpretable Image Classifier Using Deformable Prototypes
Nov 29, 2021
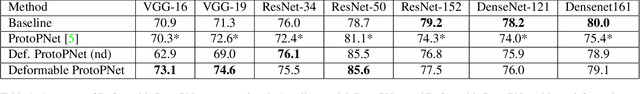
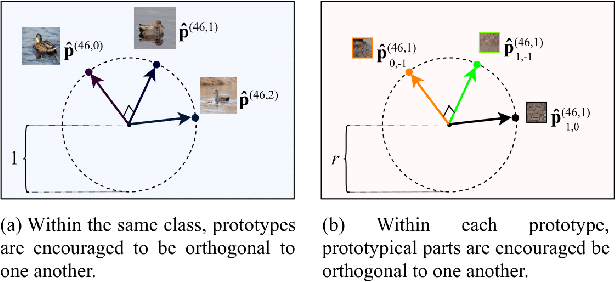
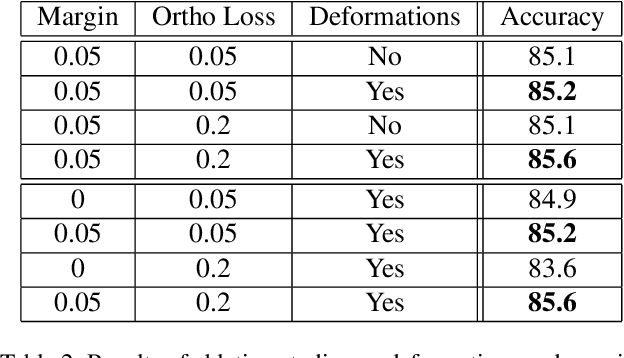
Abstract:Machine learning has been widely adopted in many domains, including high-stakes applications such as healthcare, finance, and criminal justice. To address concerns of fairness, accountability and transparency, predictions made by machine learning models in these critical domains must be interpretable. One line of work approaches this challenge by integrating the power of deep neural networks and the interpretability of case-based reasoning to produce accurate yet interpretable image classification models. These models generally classify input images by comparing them with prototypes learned during training, yielding explanations in the form of "this looks like that." However, methods from this line of work use spatially rigid prototypes, which cannot explicitly account for pose variations. In this paper, we address this shortcoming by proposing a case-based interpretable neural network that provides spatially flexible prototypes, called a deformable prototypical part network (Deformable ProtoPNet). In a Deformable ProtoPNet, each prototype is made up of several prototypical parts that adaptively change their relative spatial positions depending on the input image. This enables each prototype to detect object features with a higher tolerance to spatial transformations, as the parts within a prototype are allowed to move. Consequently, a Deformable ProtoPNet can explicitly capture pose variations, improving both model accuracy and the richness of explanations provided. Compared to other case-based interpretable models using prototypes, our approach achieves competitive accuracy, gives an explanation with greater context, and is easier to train, thus enabling wider use of interpretable models for computer vision.
Interpretable Mammographic Image Classification using Cased-Based Reasoning and Deep Learning
Jul 12, 2021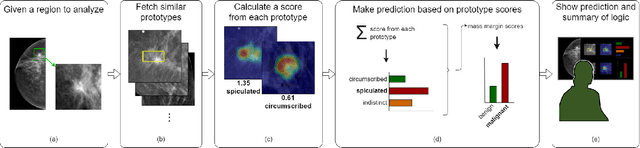
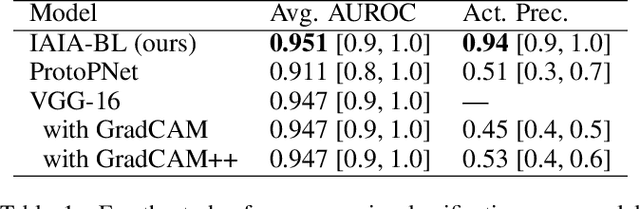
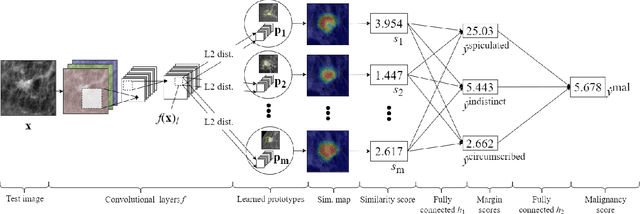
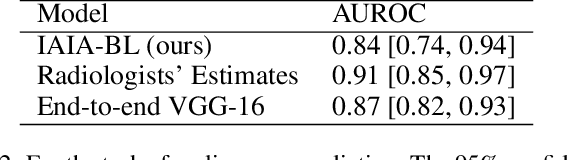
Abstract:When we deploy machine learning models in high-stakes medical settings, we must ensure these models make accurate predictions that are consistent with known medical science. Inherently interpretable networks address this need by explaining the rationale behind each decision while maintaining equal or higher accuracy compared to black-box models. In this work, we present a novel interpretable neural network algorithm that uses case-based reasoning for mammography. Designed to aid a radiologist in their decisions, our network presents both a prediction of malignancy and an explanation of that prediction using known medical features. In order to yield helpful explanations, the network is designed to mimic the reasoning processes of a radiologist: our network first detects the clinically relevant semantic features of each image by comparing each new image with a learned set of prototypical image parts from the training images, then uses those clinical features to predict malignancy. Compared to other methods, our model detects clinical features (mass margins) with equal or higher accuracy, provides a more detailed explanation of its prediction, and is better able to differentiate the classification-relevant parts of the image.
IAIA-BL: A Case-based Interpretable Deep Learning Model for Classification of Mass Lesions in Digital Mammography
Mar 23, 2021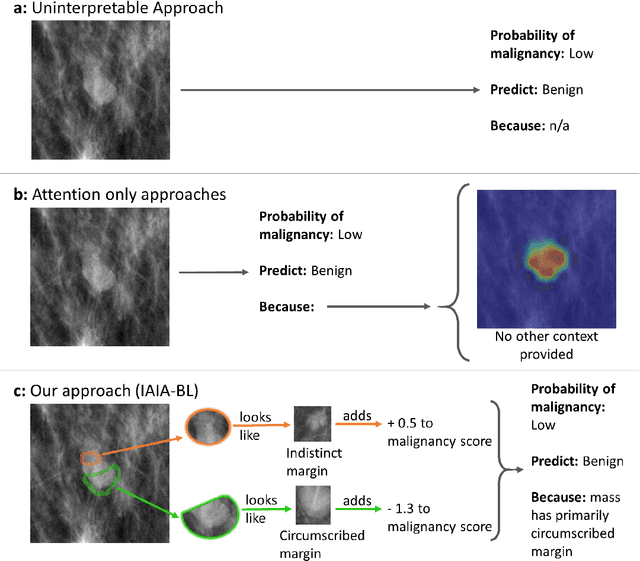
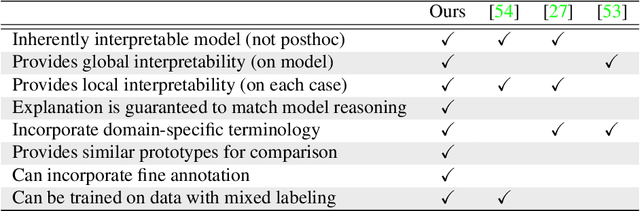
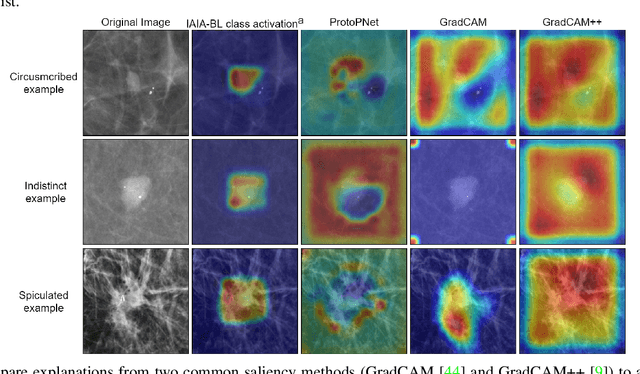
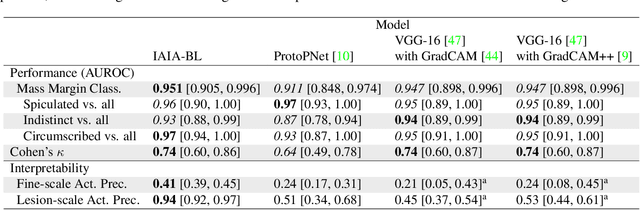
Abstract:Interpretability in machine learning models is important in high-stakes decisions, such as whether to order a biopsy based on a mammographic exam. Mammography poses important challenges that are not present in other computer vision tasks: datasets are small, confounding information is present, and it can be difficult even for a radiologist to decide between watchful waiting and biopsy based on a mammogram alone. In this work, we present a framework for interpretable machine learning-based mammography. In addition to predicting whether a lesion is malignant or benign, our work aims to follow the reasoning processes of radiologists in detecting clinically relevant semantic features of each image, such as the characteristics of the mass margins. The framework includes a novel interpretable neural network algorithm that uses case-based reasoning for mammography. Our algorithm can incorporate a combination of data with whole image labelling and data with pixel-wise annotations, leading to better accuracy and interpretability even with a small number of images. Our interpretable models are able to highlight the classification-relevant parts of the image, whereas other methods highlight healthy tissue and confounding information. Our models are decision aids, rather than decision makers, aimed at better overall human-machine collaboration. We do not observe a loss in mass margin classification accuracy over a black box neural network trained on the same data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge