Joseph Y. Lo
Tri-Reader: An Open-Access, Multi-Stage AI Pipeline for First-Pass Lung Nodule Annotation in Screening CT
Jan 27, 2026Abstract:Using multiple open-access models trained on public datasets, we developed Tri-Reader, a comprehensive, freely available pipeline that integrates lung segmentation, nodule detection, and malignancy classification into a unified tri-stage workflow. The pipeline is designed to prioritize sensitivity while reducing the candidate burden for annotators. To ensure accuracy and generalizability across diverse practices, we evaluated Tri-Reader on multiple internal and external datasets as compared with expert annotations and dataset-provided reference standards.
Organ-Aware Attention Improves CT Triage and Classification
Jan 19, 2026Abstract:There is an urgent need for triage and classification of high-volume medical imaging modalities such as computed tomography (CT), which can improve patient care and mitigate radiologist burnout. Study-level CT triage requires calibrated predictions with localized evidence; however, off-the-shelf Vision Language Models (VLM) struggle with 3D anatomy, protocol shifts, and noisy report supervision. This study used the two largest publicly available chest CT datasets: CT-RATE and RADCHEST-CT (held-out external test set). Our carefully tuned supervised baseline (instantiated as a simple Global Average Pooling head) establishes a new supervised state of the art, surpassing all reported linear-probe VLMs. Building on this baseline, we present ORACLE-CT, an encoder-agnostic, organ-aware head that pairs Organ-Masked Attention (mask-restricted, per-organ pooling that yields spatial evidence) with Organ-Scalar Fusion (lightweight fusion of normalized volume and mean-HU cues). In the chest setting, ORACLE-CT masked attention model achieves AUROC 0.86 on CT-RATE; in the abdomen setting, on MERLIN (30 findings), our supervised baseline exceeds a reproduced zero-shot VLM baseline obtained by running publicly released weights through our pipeline, and adding masked attention plus scalar fusion further improves performance to AUROC 0.85. Together, these results deliver state-of-the-art supervised classification performance across both chest and abdomen CT under a unified evaluation protocol. The source code is available at https://github.com/lavsendahal/oracle-ct.
NodMAISI: Nodule-Oriented Medical AI for Synthetic Imaging
Dec 19, 2025Abstract:Objective: Although medical imaging datasets are increasingly available, abnormal and annotation-intensive findings critical to lung cancer screening, particularly small pulmonary nodules, remain underrepresented and inconsistently curated. Methods: We introduce NodMAISI, an anatomically constrained, nodule-oriented CT synthesis and augmentation framework trained on a unified multi-source cohort (7,042 patients, 8,841 CTs, 14,444 nodules). The framework integrates: (i) a standardized curation and annotation pipeline linking each CT with organ masks and nodule-level annotations, (ii) a ControlNet-conditioned rectified-flow generator built on MAISI-v2's foundational blocks to enforce anatomy- and lesion-consistent synthesis, and (iii) lesion-aware augmentation that perturbs nodule masks (controlled shrinkage) while preserving surrounding anatomy to generate paired CT variants. Results: Across six public test datasets, NodMAISI improved distributional fidelity relative to MAISI-v2 (real-to-synthetic FID range 1.18 to 2.99 vs 1.69 to 5.21). In lesion detectability analysis using a MONAI nodule detector, NodMAISI substantially increased average sensitivity and more closely matched clinical scans (IMD-CT: 0.69 vs 0.39; DLCS24: 0.63 vs 0.20), with the largest gains for sub-centimeter nodules where MAISI-v2 frequently failed to reproduce the conditioned lesion. In downstream nodule-level malignancy classification trained on LUNA25 and externally evaluated on LUNA16, LNDbv4, and DLCS24, NodMAISI augmentation improved AUC by 0.07 to 0.11 at <=20% clinical data and by 0.12 to 0.21 at 10%, consistently narrowing the performance gap under data scarcity.
SYN-LUNGS: Towards Simulating Lung Nodules with Anatomy-Informed Digital Twins for AI Training
Feb 28, 2025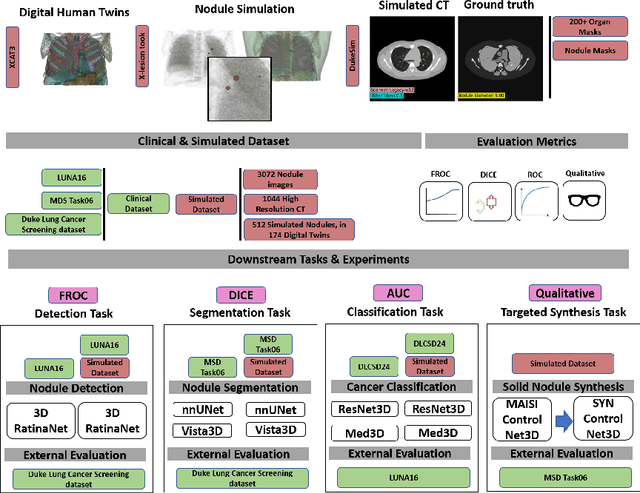
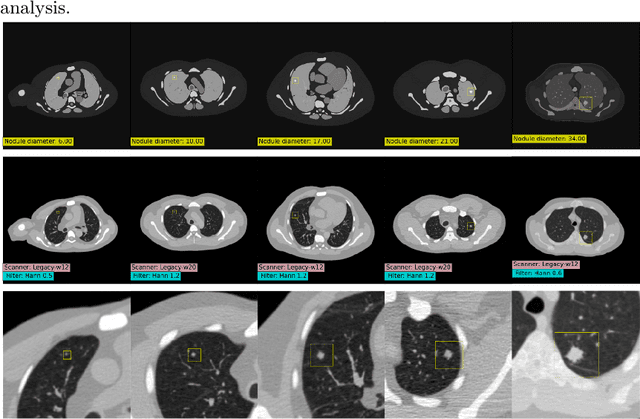
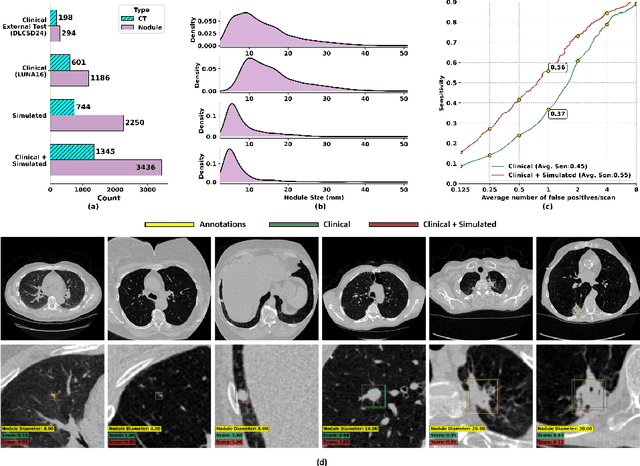
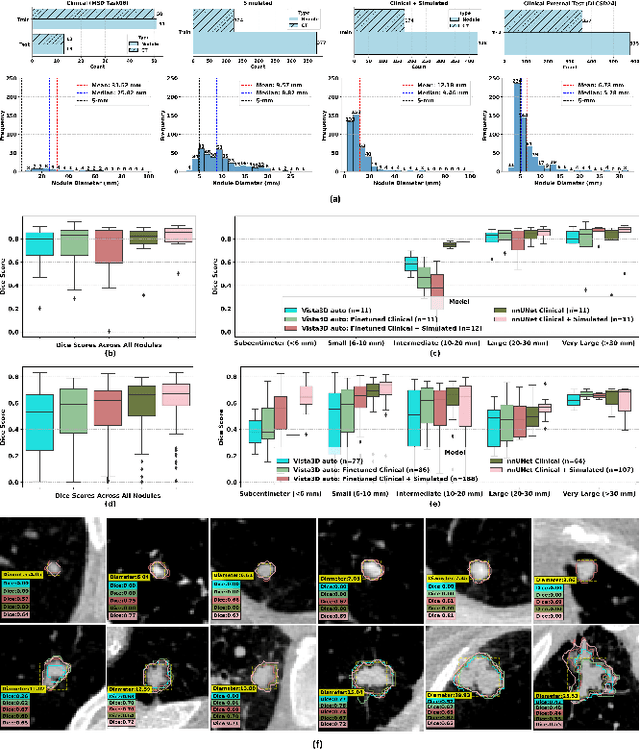
Abstract:AI models for lung cancer screening are limited by data scarcity, impacting generalizability and clinical applicability. Generative models address this issue but are constrained by training data variability. We introduce SYN-LUNGS, a framework for generating high-quality 3D CT images with detailed annotations. SYN-LUNGS integrates XCAT3 phantoms for digital twin generation, X-Lesions for nodule simulation (varying size, location, and appearance), and DukeSim for CT image formation with vendor and parameter variability. The dataset includes 3,072 nodule images from 1,044 simulated CT scans, with 512 lesions and 174 digital twins. Models trained on clinical + simulated data outperform clinical only models, achieving 10% improvement in detection, 2-9% in segmentation and classification, and enhanced synthesis.By incorporating anatomy-informed simulations, SYN-LUNGS provides a scalable approach for AI model development, particularly in rare disease representation and improving model reliability.
FPN-IAIA-BL: A Multi-Scale Interpretable Deep Learning Model for Classification of Mass Margins in Digital Mammography
Jun 10, 2024



Abstract:Digital mammography is essential to breast cancer detection, and deep learning offers promising tools for faster and more accurate mammogram analysis. In radiology and other high-stakes environments, uninterpretable ("black box") deep learning models are unsuitable and there is a call in these fields to make interpretable models. Recent work in interpretable computer vision provides transparency to these formerly black boxes by utilizing prototypes for case-based explanations, achieving high accuracy in applications including mammography. However, these models struggle with precise feature localization, reasoning on large portions of an image when only a small part is relevant. This paper addresses this gap by proposing a novel multi-scale interpretable deep learning model for mammographic mass margin classification. Our contribution not only offers an interpretable model with reasoning aligned with radiologist practices, but also provides a general architecture for computer vision with user-configurable prototypes from coarse- to fine-grained prototypes.
AI in Lung Health: Benchmarking Detection and Diagnostic Models Across Multiple CT Scan Datasets
May 07, 2024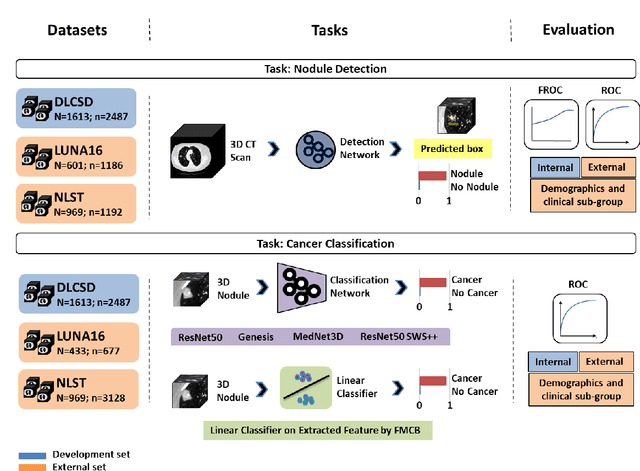
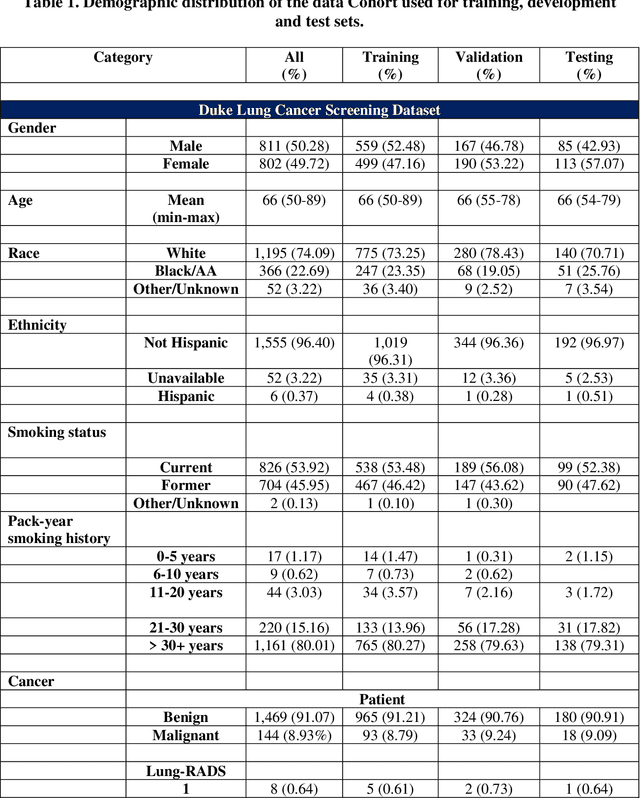
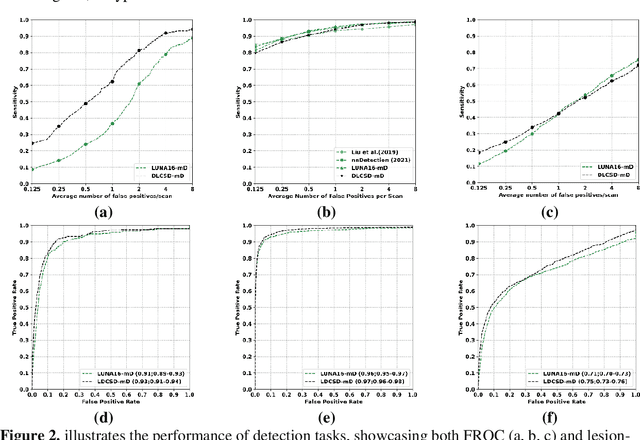
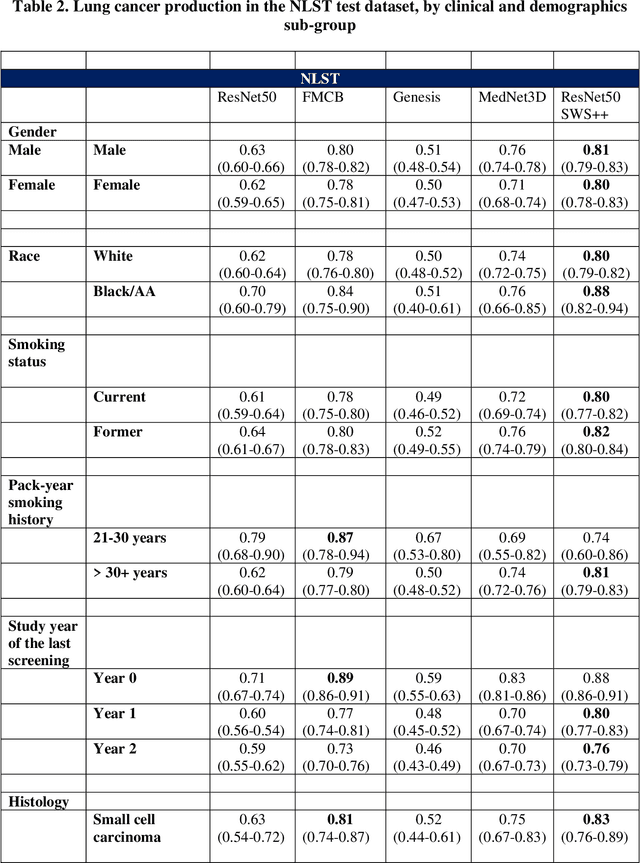
Abstract:BACKGROUND: Lung cancer's high mortality rate can be mitigated by early detection, which is increasingly reliant on artificial intelligence (AI) for diagnostic imaging. However, the performance of AI models is contingent upon the datasets used for their training and validation. METHODS: This study developed and validated the DLCSD-mD and LUNA16-mD models utilizing the Duke Lung Cancer Screening Dataset (DLCSD), encompassing over 2,000 CT scans with more than 3,000 annotations. These models were rigorously evaluated against the internal DLCSD and external LUNA16 and NLST datasets, aiming to establish a benchmark for imaging-based performance. The assessment focused on creating a standardized evaluation framework to facilitate consistent comparison with widely utilized datasets, ensuring a comprehensive validation of the model's efficacy. Diagnostic accuracy was assessed using free-response receiver operating characteristic (FROC) and area under the curve (AUC) analyses. RESULTS: On the internal DLCSD set, the DLCSD-mD model achieved an AUC of 0.93 (95% CI:0.91-0.94), demonstrating high accuracy. Its performance was sustained on the external datasets, with AUCs of 0.97 (95% CI: 0.96-0.98) on LUNA16 and 0.75 (95% CI: 0.73-0.76) on NLST. Similarly, the LUNA16-mD model recorded an AUC of 0.96 (95% CI: 0.95-0.97) on its native dataset and showed transferable diagnostic performance with AUCs of 0.91 (95% CI: 0.89-0.93) on DLCSD and 0.71 (95% CI: 0.70-0.72) on NLST. CONCLUSION: The DLCSD-mD model exhibits reliable performance across different datasets, establishing the DLCSD as a robust benchmark for lung cancer detection and diagnosis. Through the provision of our models and code to the public domain, we aim to accelerate the development of AI-based diagnostic tools and encourage reproducibility and collaborative advancements within the medical machine-learning (ML) field.
VLST: Virtual Lung Screening Trial for Lung Cancer Detection Using Virtual Imaging Trial
Apr 17, 2024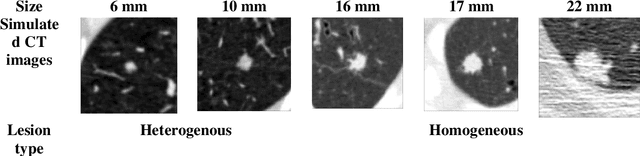
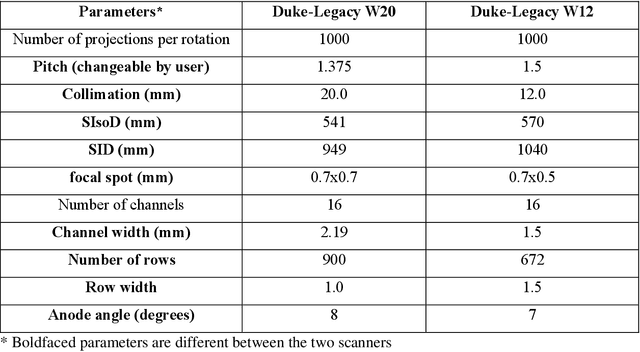
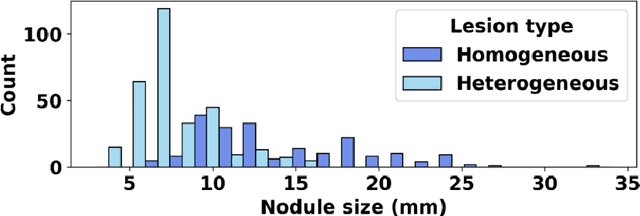
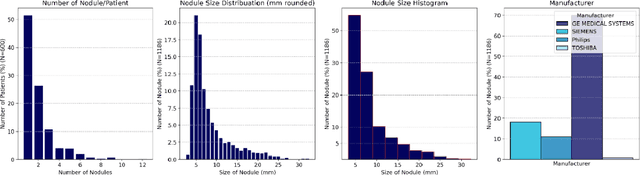
Abstract:Importance: The efficacy of lung cancer screening can be significantly impacted by the imaging modality used. This Virtual Lung Screening Trial (VLST) addresses the critical need for precision in lung cancer diagnostics and the potential for reducing unnecessary radiation exposure in clinical settings. Objectives: To establish a virtual imaging trial (VIT) platform that accurately simulates real-world lung screening trials (LSTs) to assess the diagnostic accuracy of CT and CXR modalities. Design, Setting, and Participants: Utilizing computational models and machine learning algorithms, we created a diverse virtual patient population. The cohort, designed to mirror real-world demographics, was assessed using virtual imaging techniques that reflect historical imaging technologies. Main Outcomes and Measures: The primary outcome was the difference in the Area Under the Curve (AUC) for CT and CXR modalities across lesion types and sizes. Results: The study analyzed 298 CT and 313 CXR simulated images from 313 virtual patients, with a lesion-level AUC of 0.81 (95% CI: 0.78-0.84) for CT and 0.55 (95% CI: 0.53-0.56) for CXR. At the patient level, CT demonstrated an AUC of 0.85 (95% CI: 0.80-0.89), compared to 0.53 (95% CI: 0.47-0.60) for CXR. Subgroup analyses indicated CT's superior performance in detecting homogeneous lesions (AUC of 0.97 for lesion-level) and heterogeneous lesions (AUC of 0.71 for lesion-level) as well as in identifying larger nodules (AUC of 0.98 for nodules > 8 mm). Conclusion and Relevance: The VIT platform validated the superior diagnostic accuracy of CT over CXR, especially for smaller nodules, underscoring its potential to replicate real clinical imaging trials. These findings advocate for the integration of virtual trials in the evaluation and improvement of imaging-based diagnostic tools.
What limits performance of weakly supervised deep learning for chest CT classification?
Feb 06, 2024Abstract:Weakly supervised learning with noisy data has drawn attention in the medical imaging community due to the sparsity of high-quality disease labels. However, little is known about the limitations of such weakly supervised learning and the effect of these constraints on disease classification performance. In this paper, we test the effects of such weak supervision by examining model tolerance for three conditions. First, we examined model tolerance for noisy data by incrementally increasing error in the labels within the training data. Second, we assessed the impact of dataset size by varying the amount of training data. Third, we compared performance differences between binary and multi-label classification. Results demonstrated that the model could endure up to 10% added label error before experiencing a decline in disease classification performance. Disease classification performance steadily rose as the amount of training data was increased for all disease classes, before experiencing a plateau in performance at 75% of training data. Last, the binary model outperformed the multilabel model in every disease category. However, such interpretations may be misleading, as the binary model was heavily influenced by co-occurring diseases and may not have learned the specific features of the disease in the image. In conclusion, this study may help the medical imaging community understand the benefits and risks of weak supervision with noisy labels. Such studies demonstrate the need to build diverse, large-scale datasets and to develop explainable and responsible AI.
Large Intestine 3D Shape Refinement Using Point Diffusion Models for Digital Phantom Generation
Sep 15, 2023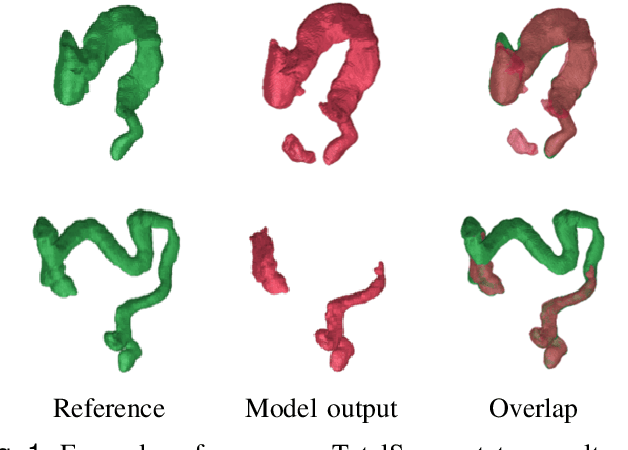
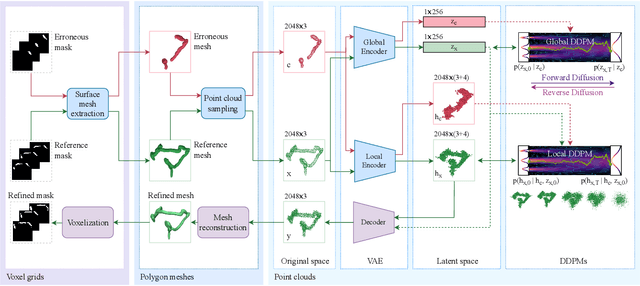
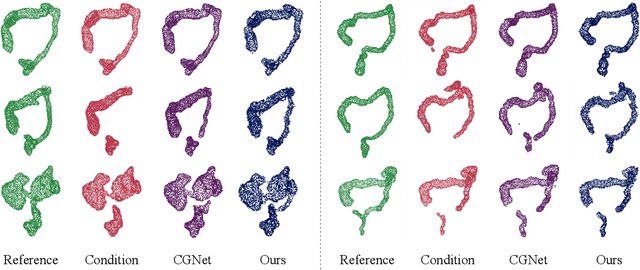
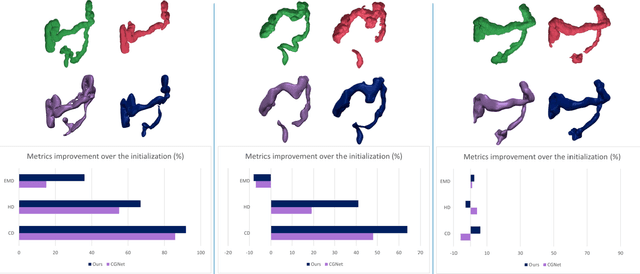
Abstract:Accurate 3D modeling of human organs plays a crucial role in building computational phantoms for virtual imaging trials. However, generating anatomically plausible reconstructions of organ surfaces from computed tomography scans remains challenging for many structures in the human body. This challenge is particularly evident when dealing with the large intestine. In this study, we leverage recent advancements in geometric deep learning and denoising diffusion probabilistic models to refine the segmentation results of the large intestine. We begin by representing the organ as point clouds sampled from the surface of the 3D segmentation mask. Subsequently, we employ a hierarchical variational autoencoder to obtain global and local latent representations of the organ's shape. We train two conditional denoising diffusion models in the hierarchical latent space to perform shape refinement. To further enhance our method, we incorporate a state-of-the-art surface reconstruction model, allowing us to generate smooth meshes from the obtained complete point clouds. Experimental results demonstrate the effectiveness of our approach in capturing both the global distribution of the organ's shape and its fine details. Our complete refinement pipeline demonstrates remarkable enhancements in surface representation compared to the initial segmentation, reducing the Chamfer distance by 70%, the Hausdorff distance by 32%, and the Earth Mover's distance by 6%. By combining geometric deep learning, denoising diffusion models, and advanced surface reconstruction techniques, our proposed method offers a promising solution for accurately modeling the large intestine's surface and can easily be extended to other anatomical structures.
Data diversity and virtual imaging in AI-based diagnosis: A case study based on COVID-19
Aug 17, 2023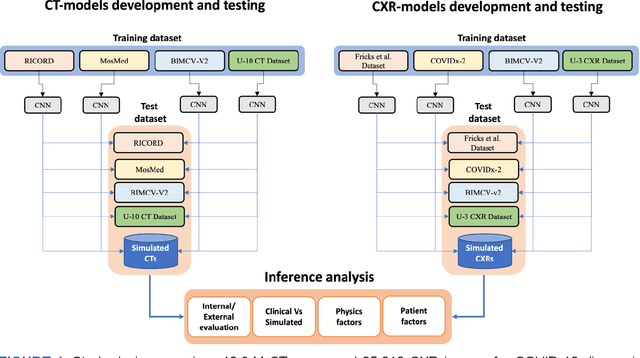
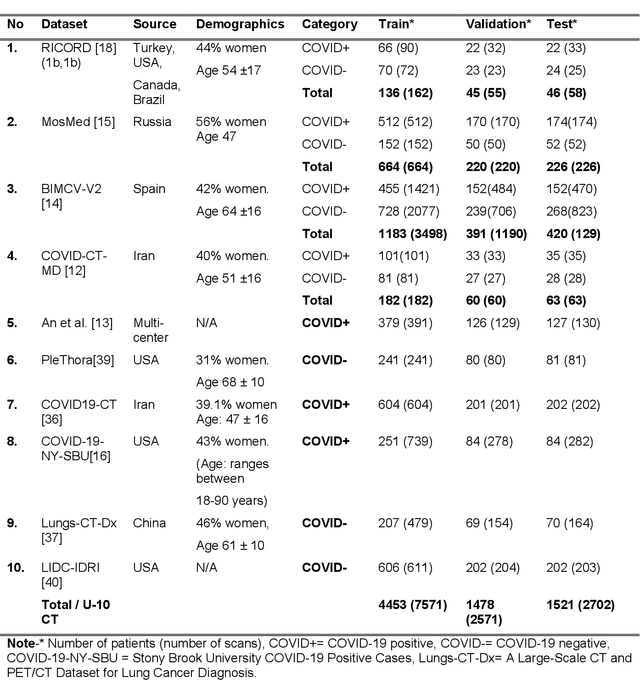
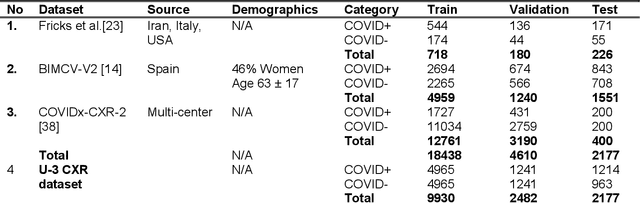
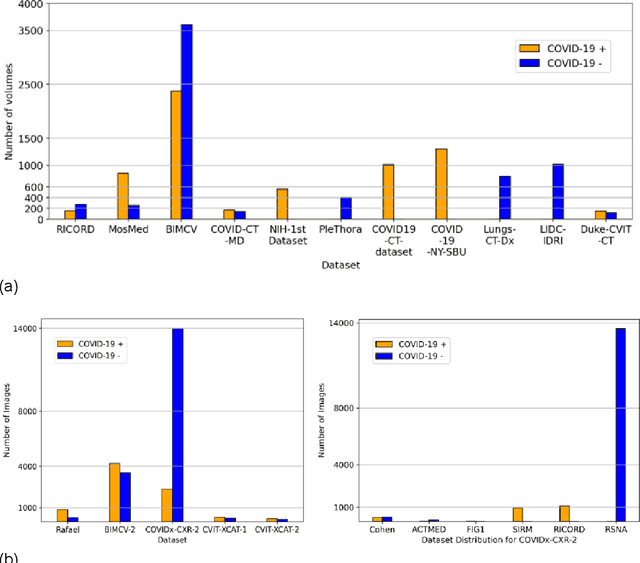
Abstract:Many studies have investigated deep-learning-based artificial intelligence (AI) models for medical imaging diagnosis of the novel coronavirus (COVID-19), with many reports of near-perfect performance. However, variability in performance and underlying data biases raise concerns about clinical generalizability. This retrospective study involved the development and evaluation of artificial intelligence (AI) models for COVID-19 diagnosis using both diverse clinical and virtually generated medical images. In addition, we conducted a virtual imaging trial to assess how AI performance is affected by several patient- and physics-based factors, including the extent of disease, radiation dose, and imaging modality of computed tomography (CT) and chest radiography (CXR). AI performance was strongly influenced by dataset characteristics including quantity, diversity, and prevalence, leading to poor generalization with up to 20% drop in receiver operating characteristic area under the curve. Model performance on virtual CT and CXR images was comparable to overall results on clinical data. Imaging dose proved to have negligible influence on the results, but the extent of the disease had a marked affect. CT results were consistently superior to those from CXR. Overall, the study highlighted the significant impact of dataset characteristics and disease extent on COVID assessment, and the relevance and potential role of virtual imaging trial techniques on developing effective evaluation of AI algorithms and facilitating translation into diagnostic practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge