Wanyi Fu
Quality or Quantity: Toward a Unified Approach for Multi-organ Segmentation in Body CT
Mar 03, 2022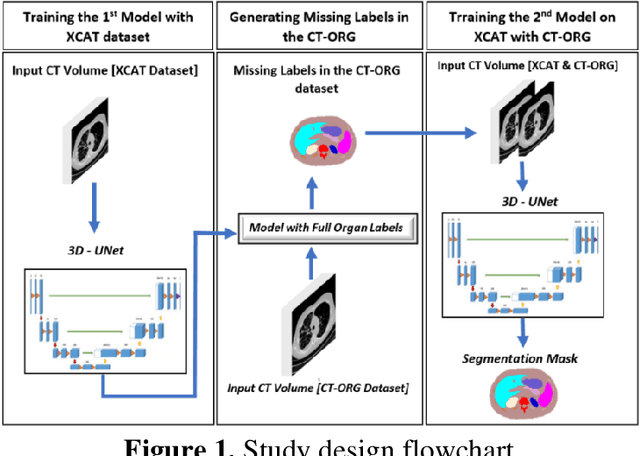
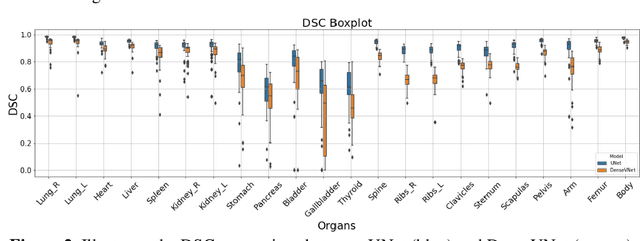
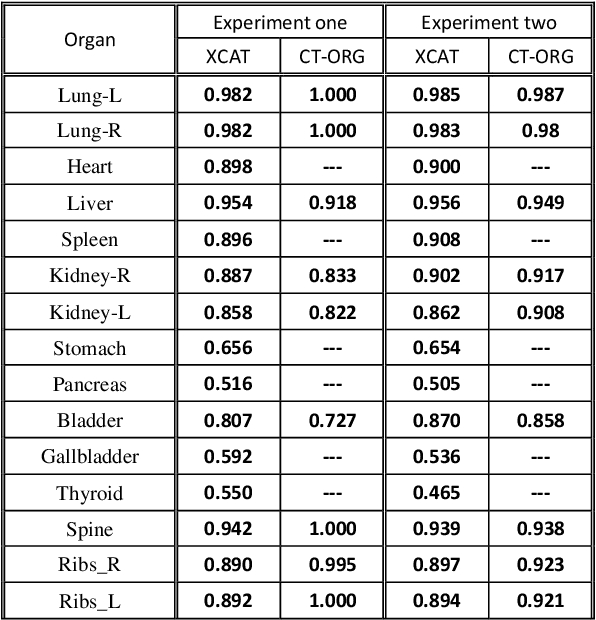
Abstract:Organ segmentation of medical images is a key step in virtual imaging trials. However, organ segmentation datasets are limited in terms of quality (because labels cover only a few organs) and quantity (since case numbers are limited). In this study, we explored the tradeoffs between quality and quantity. Our goal is to create a unified approach for multi-organ segmentation of body CT, which will facilitate the creation of large numbers of accurate virtual phantoms. Initially, we compared two segmentation architectures, 3D-Unet and DenseVNet, which were trained using XCAT data that is fully labeled with 22 organs, and chose the 3D-Unet as the better performing model. We used the XCAT-trained model to generate pseudo-labels for the CT-ORG dataset that has only 7 organs segmented. We performed two experiments: First, we trained 3D-UNet model on the XCAT dataset, representing quality data, and tested it on both XCAT and CT-ORG datasets. Second, we trained 3D-UNet after including the CT-ORG dataset into the training set to have more quantity. Performance improved for segmentation in the organs where we have true labels in both datasets and degraded when relying on pseudo-labels. When organs were labeled in both datasets, Exp-2 improved Average DSC in XCAT and CT-ORG by 1. This demonstrates that quality data is the key to improving the model's performance.
iPhantom: a framework for automated creation of individualized computational phantoms and its application to CT organ dosimetry
Aug 20, 2020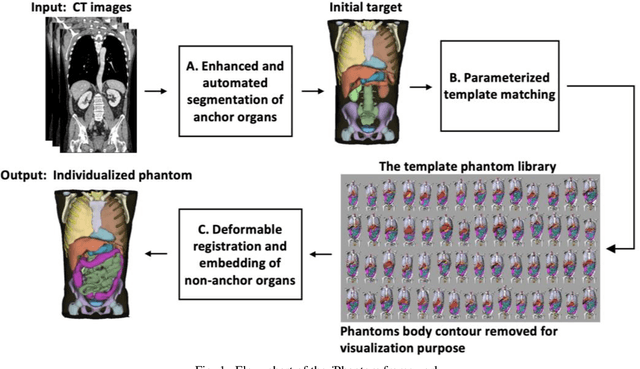
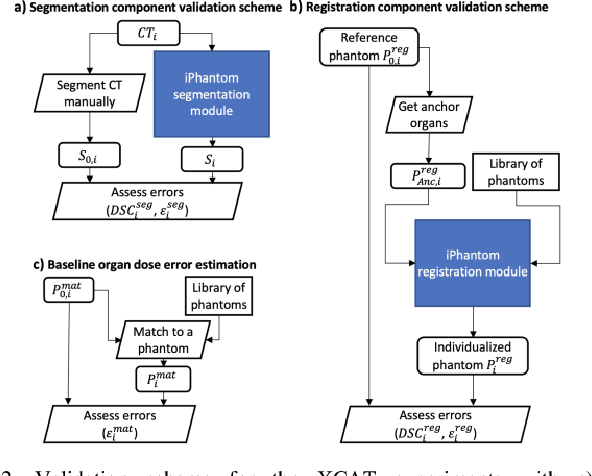
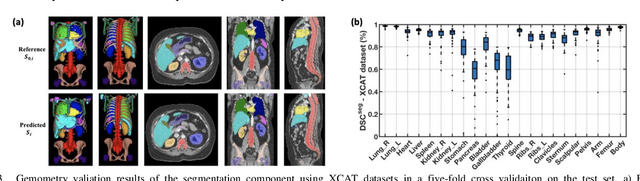
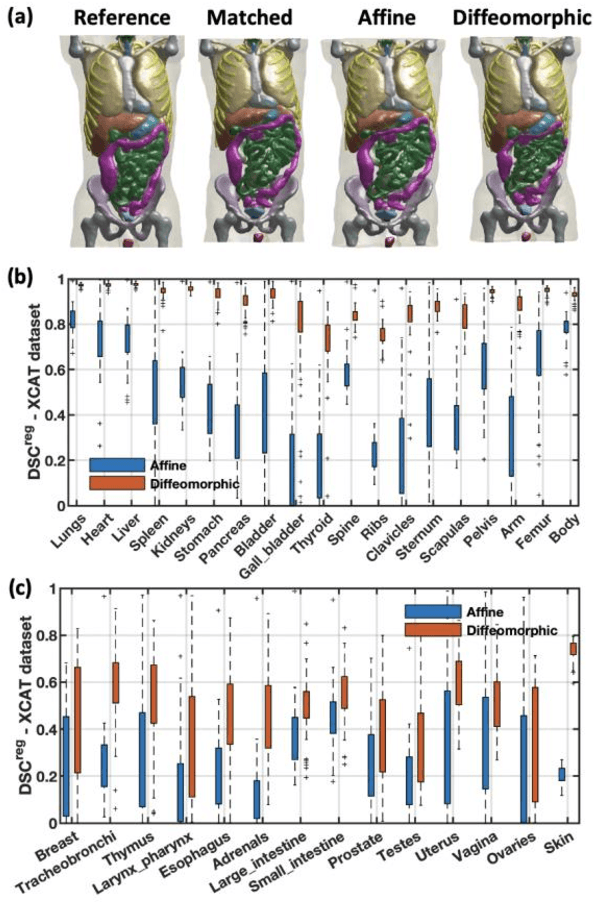
Abstract:Objective: This study aims to develop and validate a novel framework, iPhantom, for automated creation of patient-specific phantoms or digital-twins (DT) using patient medical images. The framework is applied to assess radiation dose to radiosensitive organs in CT imaging of individual patients. Method: From patient CT images, iPhantom segments selected anchor organs (e.g. liver, bones, pancreas) using a learning-based model developed for multi-organ CT segmentation. Organs challenging to segment (e.g. intestines) are incorporated from a matched phantom template, using a diffeomorphic registration model developed for multi-organ phantom-voxels. The resulting full-patient phantoms are used to assess organ doses during routine CT exams. Result: iPhantom was validated on both the XCAT (n=50) and an independent clinical (n=10) dataset with similar accuracy. iPhantom precisely predicted all organ locations with good accuracy of Dice Similarity Coefficients (DSC) >0.6 for anchor organs and DSC of 0.3-0.9 for all other organs. iPhantom showed less than 10% dose errors for the majority of organs, which was notably superior to the state-of-the-art baseline method (20-35% dose errors). Conclusion: iPhantom enables automated and accurate creation of patient-specific phantoms and, for the first time, provides sufficient and automated patient-specific dose estimates for CT dosimetry. Significance: The new framework brings the creation and application of CHPs to the level of individual CHPs through automation, achieving a wider and precise organ localization, paving the way for clinical monitoring, and personalized optimization, and large-scale research.
Weakly Supervised Multi-Organ Multi-Disease Classification of Body CT Scans
Aug 03, 2020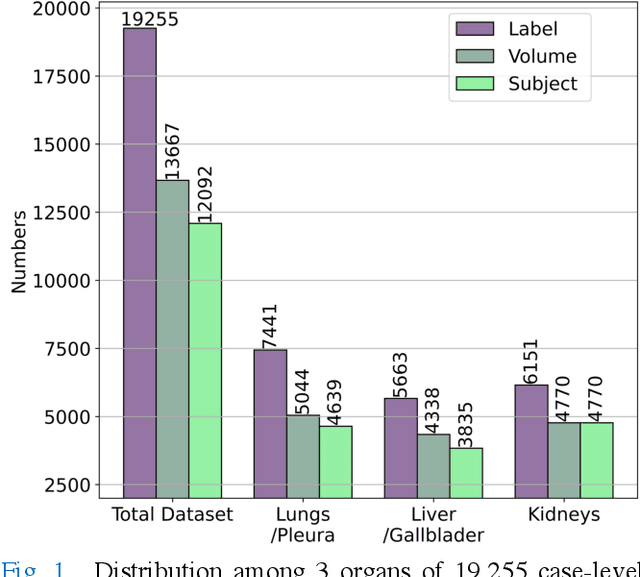
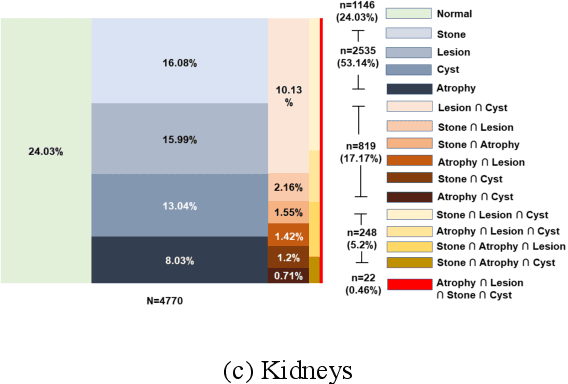
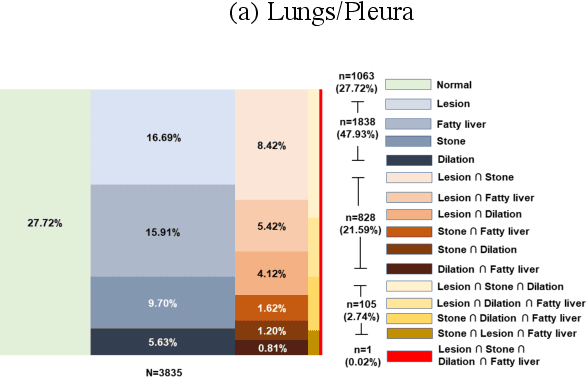
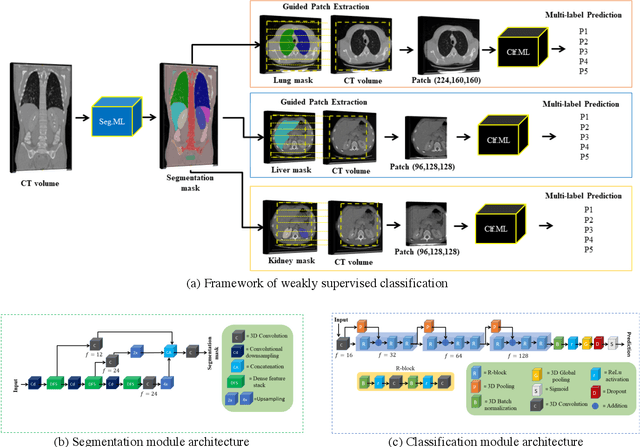
Abstract:We designed a multi-organ, multi-label disease classification algorithm for computed tomography (CT) scans using case-level labels from radiology text reports. A rule-based algorithm extracted 19,255 disease labels from reports of 13,667 body CT scans from 12,092 subjects. A 3D DenseVNet was trained to segment 3 organ systems: lungs/pleura, liver/gallbladder, and kidneys. From patches guided by segmentations, a 3D convolutional neural network provided multi-label disease classification for normality versus four common diseases per organ. The process was tested on 2,158 CT volumes with 2,875 manually obtained labels. Manual validation of the rulebased labels confirmed 91 to 99% accuracy. Results were characterized using the receiver operating characteristic area under the curve (AUC). Classification AUCs for lungs/pleura labels were as follows: atelectasis 0.77 (95% confidence intervals 0.74 to 0.81), nodule 0.65 (0.61 to 0.69), emphysema 0.89 (0.86 to 0.92), effusion 0.97 (0.96 to 0.98), and normal 0.89 (0.87 to 0.91). For liver/gallbladder, AUCs were: stone 0.62 (0.56 to 0.67), lesion 0.73 (0.69 to 0.77), dilation 0.87 (0.84 to 0.90), fatty 0.89 (0.86 to 0.92), and normal 0.82 (0.78 to 0.85). For kidneys, AUCs were: stone 0.83 (0.79 to 0.87), atrophy 0.92 (0.89 to 0.94), lesion 0.68 (0.64 to 0.72), cyst 0.70 (0.66 to 0.73), and normal 0.79 (0.75 to 0.83). In conclusion, by using automated extraction of disease labels from radiology reports, we created a weakly supervised, multi-organ, multi-disease classifier that can be easily adapted to efficiently leverage massive amounts of unannotated data associated with medical images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge