Alexandros-Stavros Iliopoulos
Communication-Efficient Graph Neural Networks with Probabilistic Neighborhood Expansion Analysis and Caching
May 04, 2023Abstract:Training and inference with graph neural networks (GNNs) on massive graphs has been actively studied since the inception of GNNs, owing to the widespread use and success of GNNs in applications such as recommendation systems and financial forensics. This paper is concerned with minibatch training and inference with GNNs that employ node-wise sampling in distributed settings, where the necessary partitioning of vertex features across distributed storage causes feature communication to become a major bottleneck that hampers scalability. To significantly reduce the communication volume without compromising prediction accuracy, we propose a policy for caching data associated with frequently accessed vertices in remote partitions. The proposed policy is based on an analysis of vertex-wise inclusion probabilities (VIP) during multi-hop neighborhood sampling, which may expand the neighborhood far beyond the partition boundaries of the graph. VIP analysis not only enables the elimination of the communication bottleneck, but it also offers a means to organize in-memory data by prioritizing GPU storage for the most frequently accessed vertex features. We present SALIENT++, which extends the prior state-of-the-art SALIENT system to work with partitioned feature data and leverages the VIP-driven caching policy. SALIENT++ retains the local training efficiency and scalability of SALIENT by using a deep pipeline and drastically reducing communication volume while consuming only a fraction of the storage required by SALIENT. We provide experimental results with the Open Graph Benchmark data sets and demonstrate that training a 3-layer GraphSAGE model with SALIENT++ on 8 single-GPU machines is 7.1 faster than with SALIENT on 1 single-GPU machine, and 12.7 faster than with DistDGL on 8 single-GPU machines.
Accelerating Training and Inference of Graph Neural Networks with Fast Sampling and Pipelining
Oct 16, 2021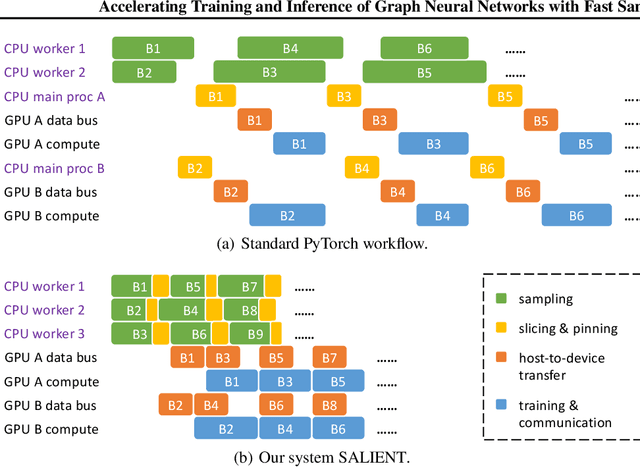
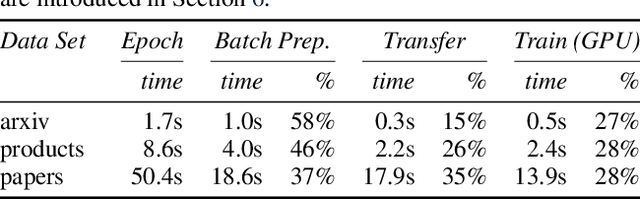
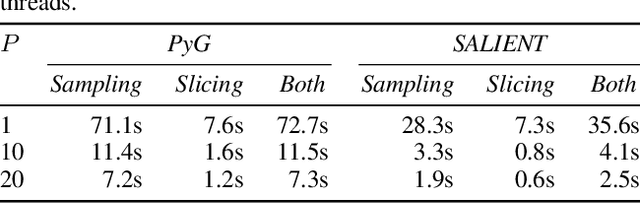
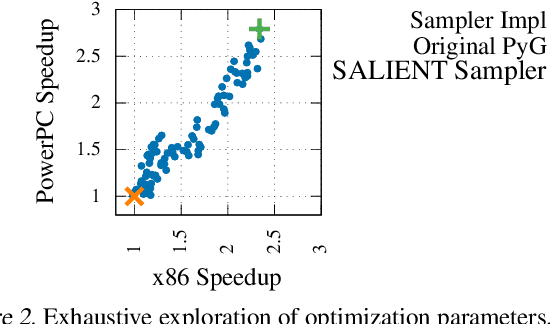
Abstract:Improving the training and inference performance of graph neural networks (GNNs) is faced with a challenge uncommon in general neural networks: creating mini-batches requires a lot of computation and data movement due to the exponential growth of multi-hop graph neighborhoods along network layers. Such a unique challenge gives rise to a diverse set of system design choices. We argue in favor of performing mini-batch training with neighborhood sampling in a distributed multi-GPU environment, under which we identify major performance bottlenecks hitherto under-explored by developers: mini-batch preparation and transfer. We present a sequence of improvements to mitigate these bottlenecks, including a performance-engineered neighborhood sampler, a shared-memory parallelization strategy, and the pipelining of batch transfer with GPU computation. We also conduct an empirical analysis that supports the use of sampling for inference, showing that test accuracies are not materially compromised. Such an observation unifies training and inference, simplifying model implementation. We report comprehensive experimental results with several benchmark data sets and GNN architectures, including a demonstration that, for the ogbn-papers100M data set, our system SALIENT achieves a speedup of 3x over a standard PyTorch-Geometric implementation with a single GPU and a further 8x parallel speedup with 16 GPUs. Therein, training a 3-layer GraphSAGE model with sampling fanout (15, 10, 5) takes 2.0 seconds per epoch and inference with fanout (20, 20, 20) takes 2.4 seconds, attaining test accuracy 64.58%.
iPhantom: a framework for automated creation of individualized computational phantoms and its application to CT organ dosimetry
Aug 20, 2020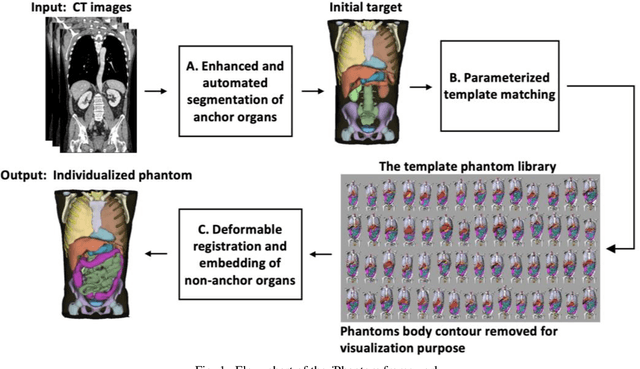
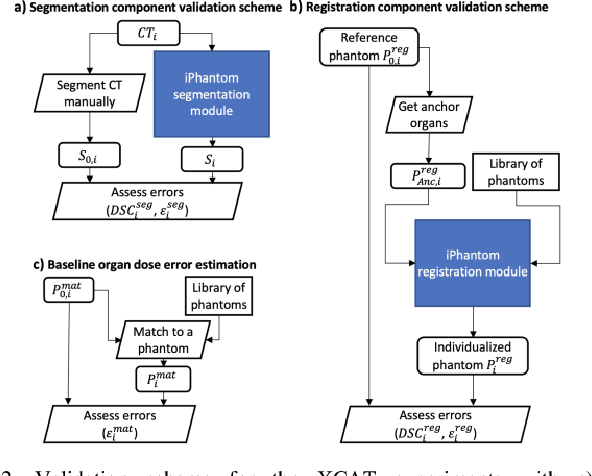
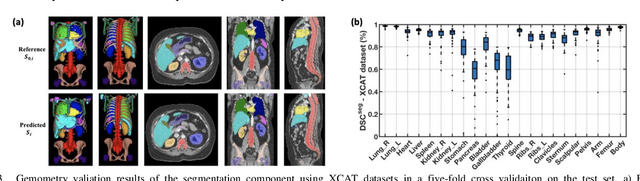
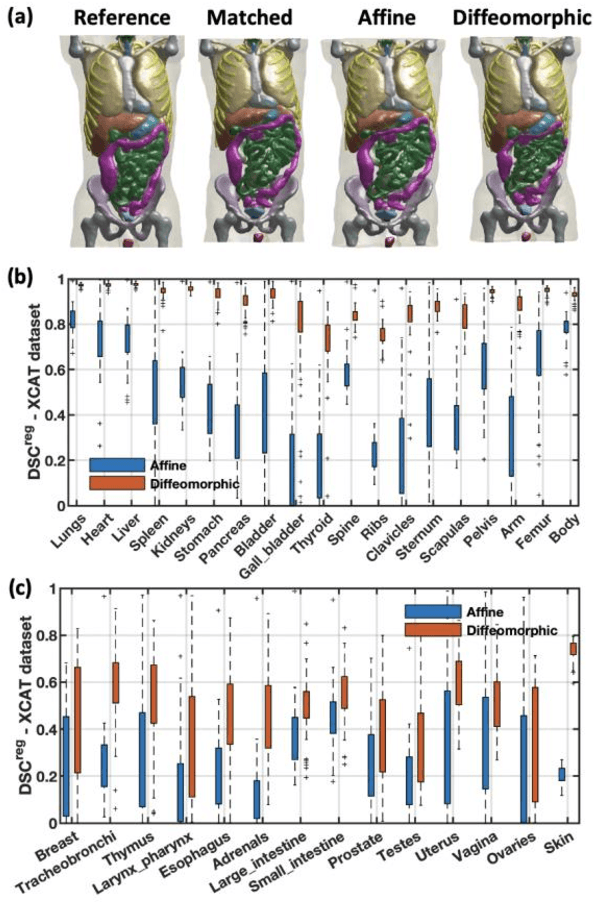
Abstract:Objective: This study aims to develop and validate a novel framework, iPhantom, for automated creation of patient-specific phantoms or digital-twins (DT) using patient medical images. The framework is applied to assess radiation dose to radiosensitive organs in CT imaging of individual patients. Method: From patient CT images, iPhantom segments selected anchor organs (e.g. liver, bones, pancreas) using a learning-based model developed for multi-organ CT segmentation. Organs challenging to segment (e.g. intestines) are incorporated from a matched phantom template, using a diffeomorphic registration model developed for multi-organ phantom-voxels. The resulting full-patient phantoms are used to assess organ doses during routine CT exams. Result: iPhantom was validated on both the XCAT (n=50) and an independent clinical (n=10) dataset with similar accuracy. iPhantom precisely predicted all organ locations with good accuracy of Dice Similarity Coefficients (DSC) >0.6 for anchor organs and DSC of 0.3-0.9 for all other organs. iPhantom showed less than 10% dose errors for the majority of organs, which was notably superior to the state-of-the-art baseline method (20-35% dose errors). Conclusion: iPhantom enables automated and accurate creation of patient-specific phantoms and, for the first time, provides sufficient and automated patient-specific dose estimates for CT dosimetry. Significance: The new framework brings the creation and application of CHPs to the level of individual CHPs through automation, achieving a wider and precise organ localization, paving the way for clinical monitoring, and personalized optimization, and large-scale research.
Spaceland Embedding of Sparse Stochastic Graphs
Jun 13, 2019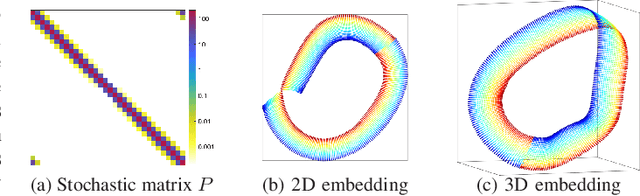
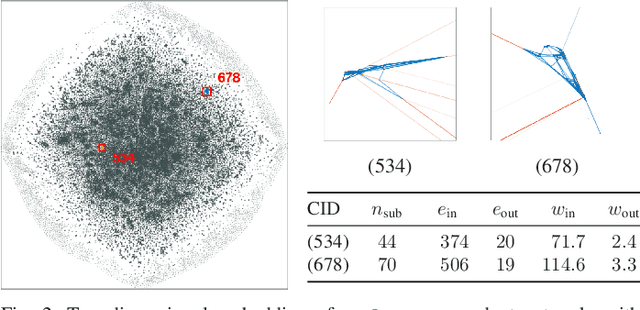
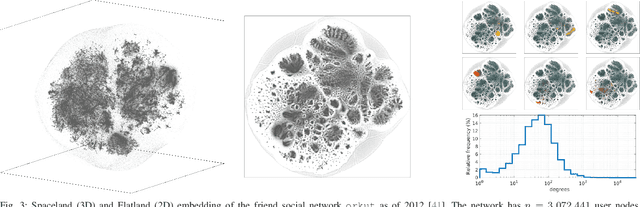
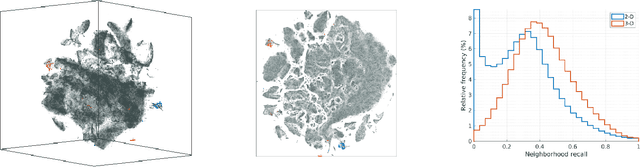
Abstract:We introduce a nonlinear method for directly embedding large, sparse, stochastic graphs into low-dimensional spaces, without requiring vertex features to reside in, or be transformed into, a metric space. Graph data and models are prevalent in real-world applications. Direct graph embedding is fundamental to many graph analysis tasks, in addition to graph visualization. We name the novel approach SG-t-SNE, as it is inspired by and builds upon the core principle of t-SNE, a widely used method for nonlinear dimensionality reduction and data visualization. We also introduce t-SNE-$\Pi$, a high-performance software for 2D, 3D embedding of large sparse graphs on personal computers with superior efficiency. It empowers SG-t-SNE with modern computing techniques for exploiting in tandem both matrix structures and memory architectures. We present elucidating embedding results on one synthetic graph and four real-world networks.
Iterative Inversion of Deformation Vector Fields with Feedback Control
Mar 28, 2018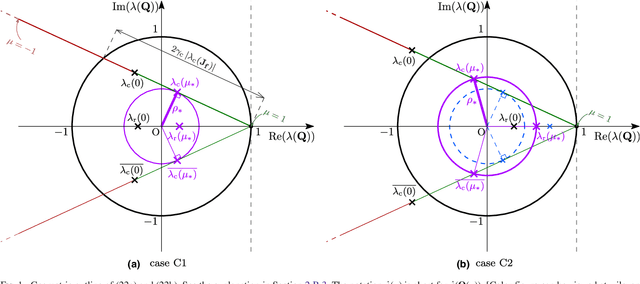
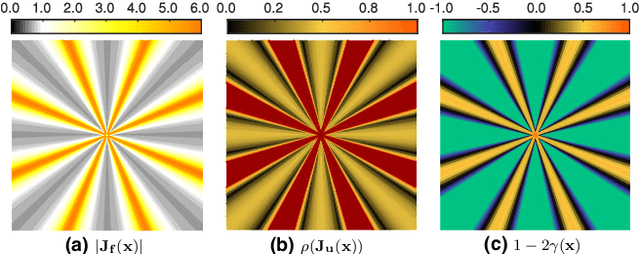
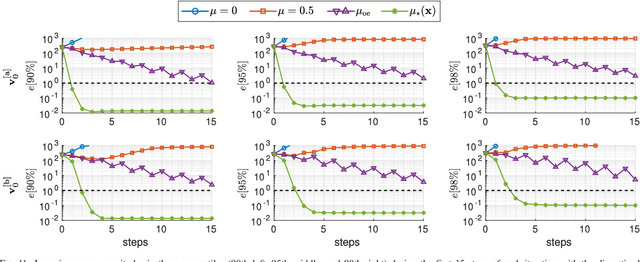
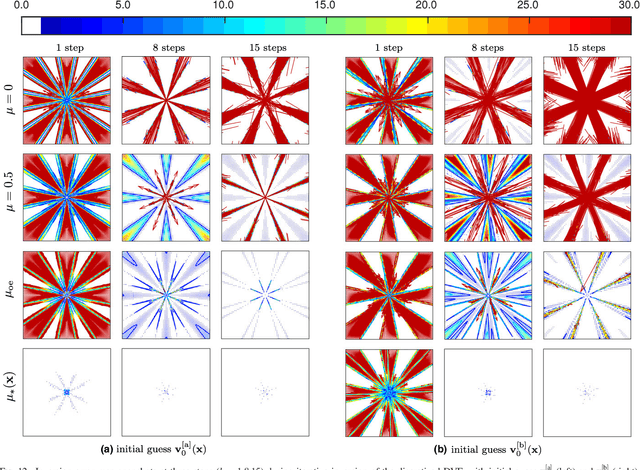
Abstract:Purpose: Often, the inverse deformation vector field (DVF) is needed together with the corresponding forward DVF in 4D reconstruction and dose calculation, adaptive radiation therapy, and simultaneous deformable registration. This study aims at improving both accuracy and efficiency of iterative algorithms for DVF inversion, and advancing our understanding of divergence and latency conditions. Method: We introduce a framework of fixed-point iteration algorithms with active feedback control for DVF inversion. Based on rigorous convergence analysis, we design control mechanisms for modulating the inverse consistency (IC) residual of the current iterate, to be used as feedback into the next iterate. The control is designed adaptively to the input DVF with the objective to enlarge the convergence area and expedite convergence. Three particular settings of feedback control are introduced: constant value over the domain throughout the iteration; alternating values between iteration steps; and spatially variant values. We also introduce three spectral measures of the displacement Jacobian for characterizing a DVF. These measures reveal the critical role of what we term the non-translational displacement component (NTDC) of the DVF. We carry out inversion experiments with an analytical DVF pair, and with DVFs associated with thoracic CT images of 6 patients at end of expiration and end of inspiration. Results: NTDC-adaptive iterations are shown to attain a larger convergence region at a faster pace compared to previous non-adaptive DVF inversion iteration algorithms. By our numerical experiments, alternating control yields smaller IC residuals and inversion errors than constant control. Spatially variant control renders smaller residuals and errors by at least an order of magnitude, compared to other schemes, in no more than 10 steps. Inversion results also show remarkable quantitative agreement with analysis-based predictions. Conclusion: Our analysis captures properties of DVF data associated with clinical CT images, and provides new understanding of iterative DVF inversion algorithms with a simple residual feedback control. Adaptive control is necessary and highly effective in the presence of non-small NTDCs. The adaptive iterations or the spectral measures, or both, may potentially be incorporated into deformable image registration methods.
Rapid Near-Neighbor Interaction of High-dimensional Data via Hierarchical Clustering
Sep 12, 2017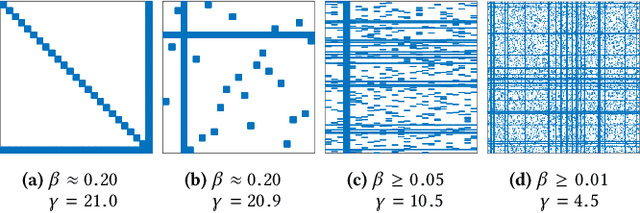
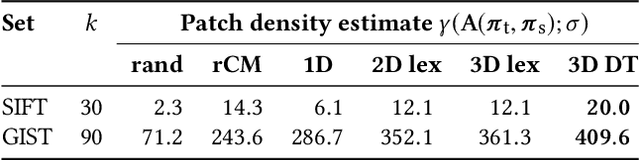
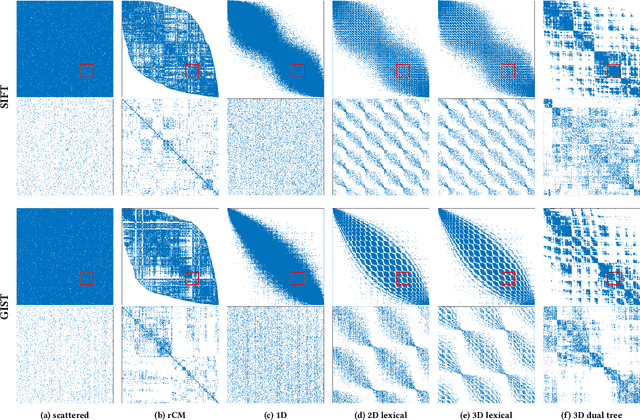
Abstract:Calculation of near-neighbor interactions among high dimensional, irregularly distributed data points is a fundamental task to many graph-based or kernel-based machine learning algorithms and applications. Such calculations, involving large, sparse interaction matrices, expose the limitation of conventional data-and-computation reordering techniques for improving space and time locality on modern computer memory hierarchies. We introduce a novel method for obtaining a matrix permutation that renders a desirable sparsity profile. The method is distinguished by the guiding principle to obtain a profile that is block-sparse with dense blocks. Our profile model and measure capture the essential properties affecting space and time locality, and permit variation in sparsity profile without imposing a restriction to a fixed pattern. The second distinction lies in an efficient algorithm for obtaining a desirable profile, via exploring and exploiting multi-scale cluster structure hidden in but intrinsic to the data. The algorithm accomplishes its task with key components for lower-dimensional embedding with data-specific principal feature axes, hierarchical data clustering, multi-level matrix compression storage, and multi-level interaction computations. We provide experimental results from case studies with two important data analysis algorithms. The resulting performance is remarkably comparable to the BLAS performance for the best-case interaction governed by a regularly banded matrix with the same sparsity.
Hyperspectral Image Classification and Clutter Detection via Multiple Structural Embeddings and Dimension Reductions
Jun 03, 2015
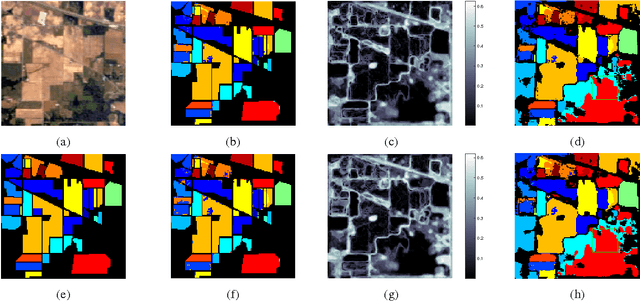
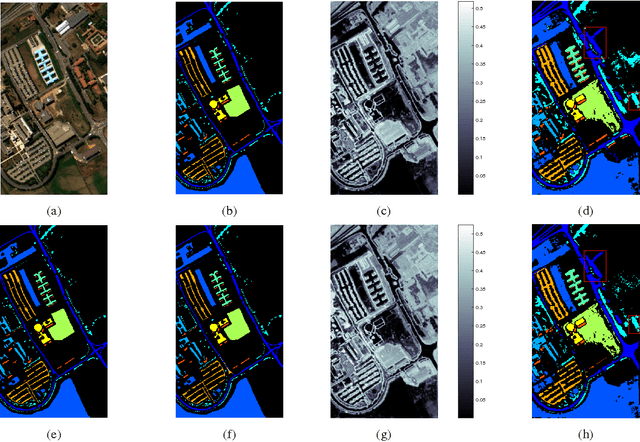
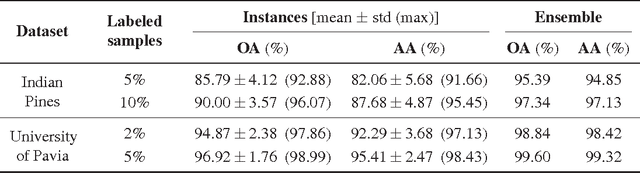
Abstract:We present a new and effective approach for Hyperspectral Image (HSI) classification and clutter detection, overcoming a few long-standing challenges presented by HSI data characteristics. Residing in a high-dimensional spectral attribute space, HSI data samples are known to be strongly correlated in their spectral signatures, exhibit nonlinear structure due to several physical laws, and contain uncertainty and noise from multiple sources. In the presented approach, we generate an adaptive, structurally enriched representation environment, and employ the locally linear embedding (LLE) in it. There are two structure layers external to LLE. One is feature space embedding: the HSI data attributes are embedded into a discriminatory feature space where spatio-spectral coherence and distinctive structures are distilled and exploited to mitigate various difficulties encountered in the native hyperspectral attribute space. The other structure layer encloses the ranges of algorithmic parameters for LLE and feature embedding, and supports a multiplexing and integrating scheme for contending with multi-source uncertainty. Experiments on two commonly used HSI datasets with a small number of learning samples have rendered remarkably high-accuracy classification results, as well as distinctive maps of detected clutter regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge