Rapid Near-Neighbor Interaction of High-dimensional Data via Hierarchical Clustering
Paper and Code
Sep 12, 2017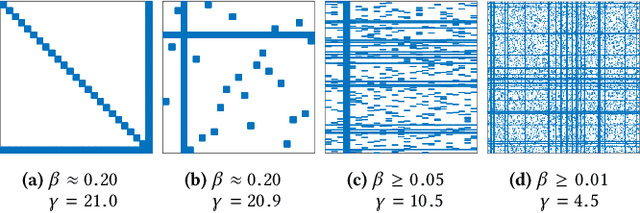
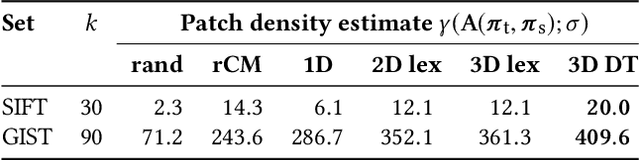
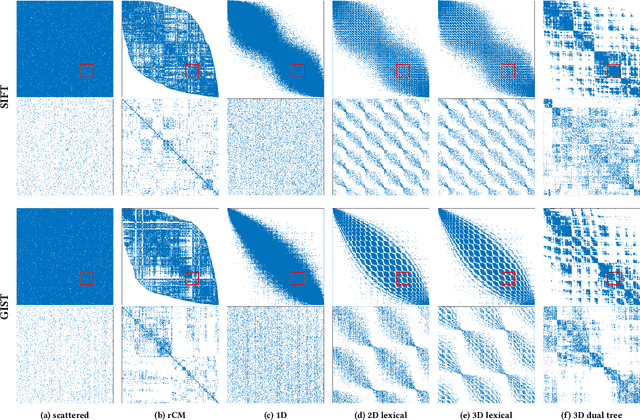
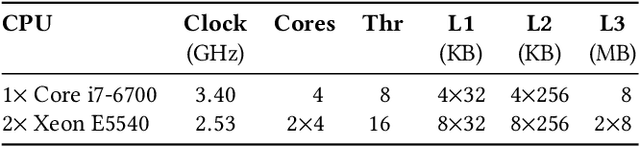
Calculation of near-neighbor interactions among high dimensional, irregularly distributed data points is a fundamental task to many graph-based or kernel-based machine learning algorithms and applications. Such calculations, involving large, sparse interaction matrices, expose the limitation of conventional data-and-computation reordering techniques for improving space and time locality on modern computer memory hierarchies. We introduce a novel method for obtaining a matrix permutation that renders a desirable sparsity profile. The method is distinguished by the guiding principle to obtain a profile that is block-sparse with dense blocks. Our profile model and measure capture the essential properties affecting space and time locality, and permit variation in sparsity profile without imposing a restriction to a fixed pattern. The second distinction lies in an efficient algorithm for obtaining a desirable profile, via exploring and exploiting multi-scale cluster structure hidden in but intrinsic to the data. The algorithm accomplishes its task with key components for lower-dimensional embedding with data-specific principal feature axes, hierarchical data clustering, multi-level matrix compression storage, and multi-level interaction computations. We provide experimental results from case studies with two important data analysis algorithms. The resulting performance is remarkably comparable to the BLAS performance for the best-case interaction governed by a regularly banded matrix with the same sparsity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge