Ehsan Samei
NodMAISI: Nodule-Oriented Medical AI for Synthetic Imaging
Dec 19, 2025Abstract:Objective: Although medical imaging datasets are increasingly available, abnormal and annotation-intensive findings critical to lung cancer screening, particularly small pulmonary nodules, remain underrepresented and inconsistently curated. Methods: We introduce NodMAISI, an anatomically constrained, nodule-oriented CT synthesis and augmentation framework trained on a unified multi-source cohort (7,042 patients, 8,841 CTs, 14,444 nodules). The framework integrates: (i) a standardized curation and annotation pipeline linking each CT with organ masks and nodule-level annotations, (ii) a ControlNet-conditioned rectified-flow generator built on MAISI-v2's foundational blocks to enforce anatomy- and lesion-consistent synthesis, and (iii) lesion-aware augmentation that perturbs nodule masks (controlled shrinkage) while preserving surrounding anatomy to generate paired CT variants. Results: Across six public test datasets, NodMAISI improved distributional fidelity relative to MAISI-v2 (real-to-synthetic FID range 1.18 to 2.99 vs 1.69 to 5.21). In lesion detectability analysis using a MONAI nodule detector, NodMAISI substantially increased average sensitivity and more closely matched clinical scans (IMD-CT: 0.69 vs 0.39; DLCS24: 0.63 vs 0.20), with the largest gains for sub-centimeter nodules where MAISI-v2 frequently failed to reproduce the conditioned lesion. In downstream nodule-level malignancy classification trained on LUNA25 and externally evaluated on LUNA16, LNDbv4, and DLCS24, NodMAISI augmentation improved AUC by 0.07 to 0.11 at <=20% clinical data and by 0.12 to 0.21 at 10%, consistently narrowing the performance gap under data scarcity.
SYN-LUNGS: Towards Simulating Lung Nodules with Anatomy-Informed Digital Twins for AI Training
Feb 28, 2025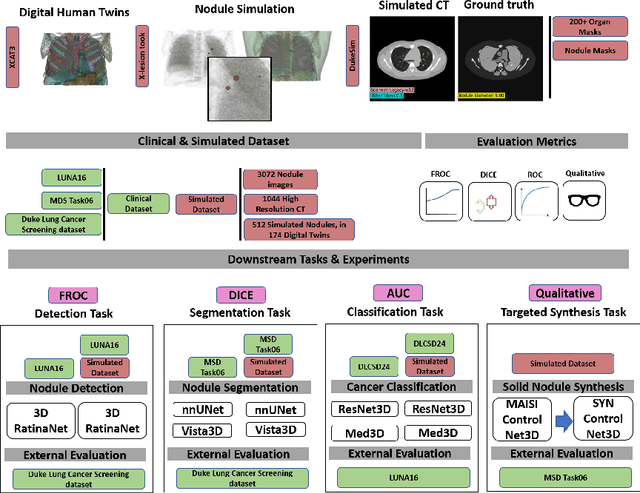
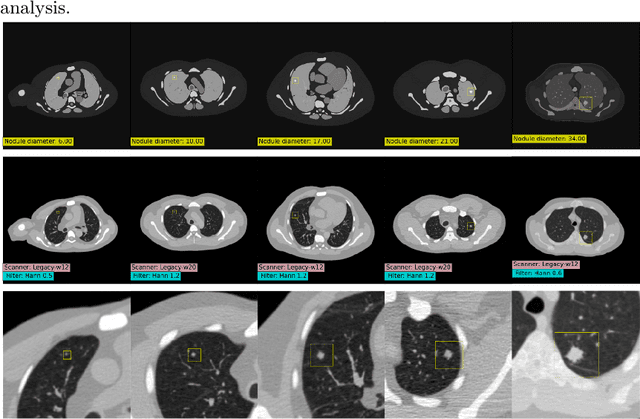
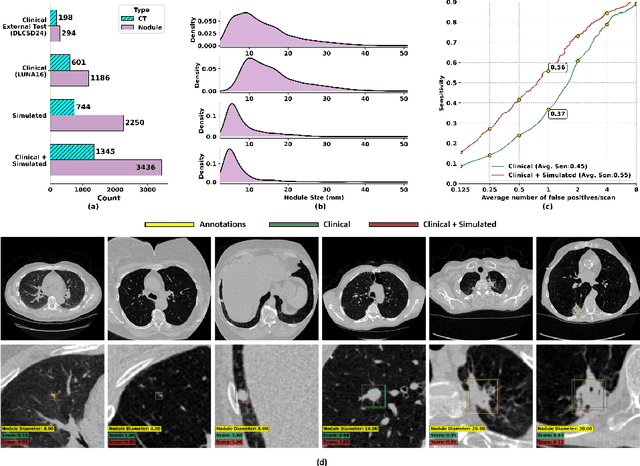
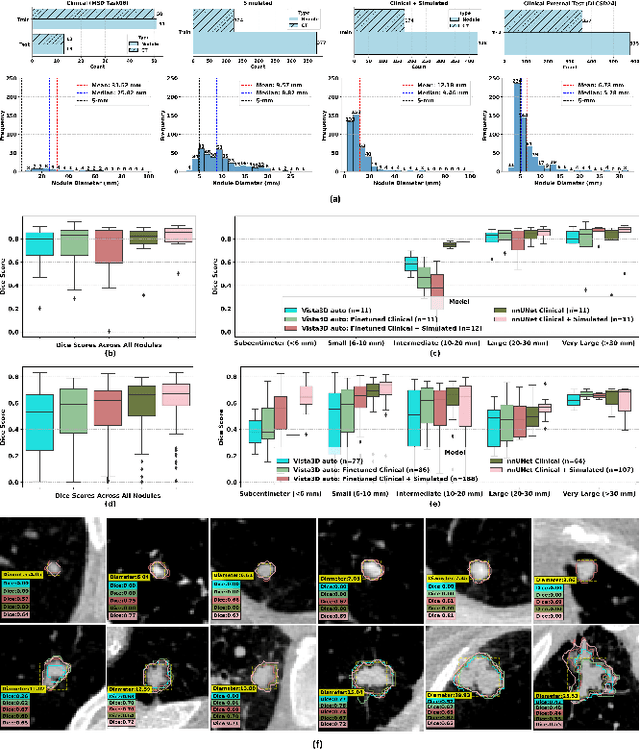
Abstract:AI models for lung cancer screening are limited by data scarcity, impacting generalizability and clinical applicability. Generative models address this issue but are constrained by training data variability. We introduce SYN-LUNGS, a framework for generating high-quality 3D CT images with detailed annotations. SYN-LUNGS integrates XCAT3 phantoms for digital twin generation, X-Lesions for nodule simulation (varying size, location, and appearance), and DukeSim for CT image formation with vendor and parameter variability. The dataset includes 3,072 nodule images from 1,044 simulated CT scans, with 512 lesions and 174 digital twins. Models trained on clinical + simulated data outperform clinical only models, achieving 10% improvement in detection, 2-9% in segmentation and classification, and enhanced synthesis.By incorporating anatomy-informed simulations, SYN-LUNGS provides a scalable approach for AI model development, particularly in rare disease representation and improving model reliability.
XCAT-2.0: A Comprehensive Library of Personalized Digital Twins Derived from CT Scans
May 18, 2024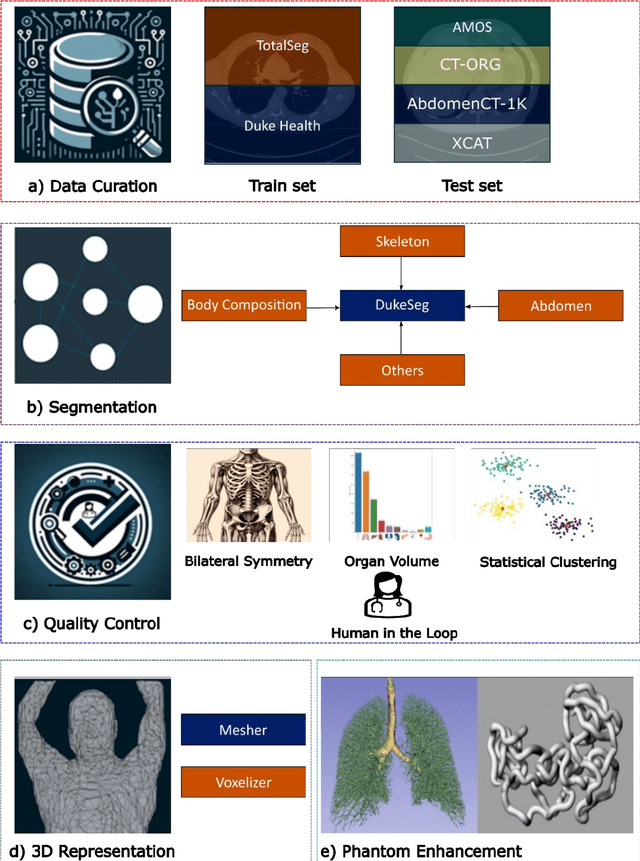
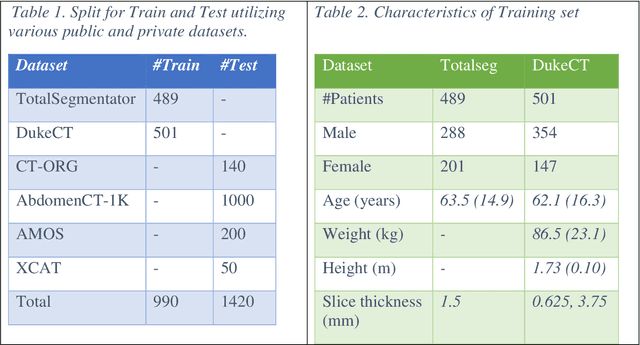
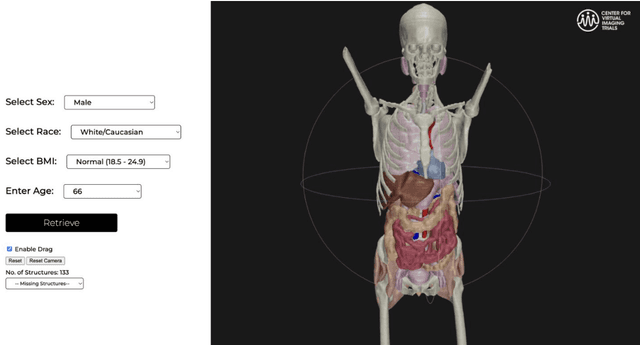
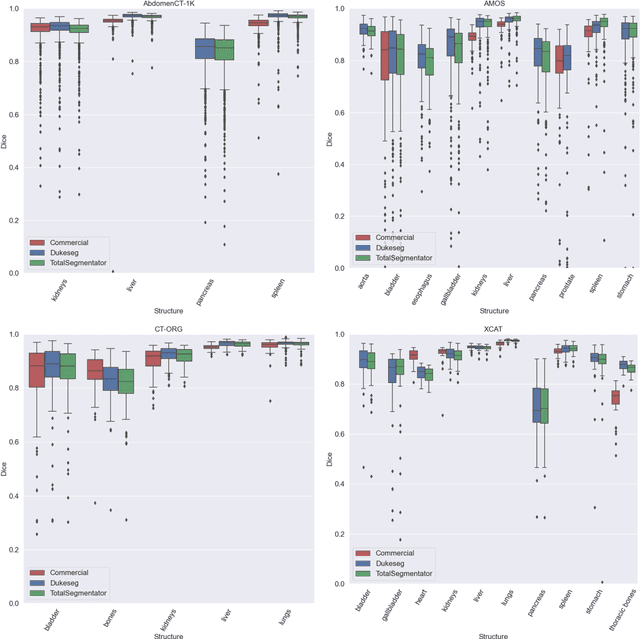
Abstract:Virtual Imaging Trials (VIT) offer a cost-effective and scalable approach for evaluating medical imaging technologies. Computational phantoms, which mimic real patient anatomy and physiology, play a central role in VIT. However, the current libraries of computational phantoms face limitations, particularly in terms of sample size and diversity. Insufficient representation of the population hampers accurate assessment of imaging technologies across different patient groups. Traditionally, phantoms were created by manual segmentation, which is a laborious and time-consuming task, impeding the expansion of phantom libraries. This study presents a framework for realistic computational phantom modeling using a suite of four deep learning segmentation models, followed by three forms of automated organ segmentation quality control. Over 2500 computational phantoms with up to 140 structures illustrating a sophisticated approach to detailed anatomical modeling are released. Phantoms are available in both voxelized and surface mesh formats. The framework is aggregated with an in-house CT scanner simulator to produce realistic CT images. The framework can potentially advance virtual imaging trials, facilitating comprehensive and reliable evaluations of medical imaging technologies. Phantoms may be requested at https://cvit.duke.edu/resources/, code, model weights, and sample CT images are available at https://xcat-2.github.io.
VLST: Virtual Lung Screening Trial for Lung Cancer Detection Using Virtual Imaging Trial
Apr 17, 2024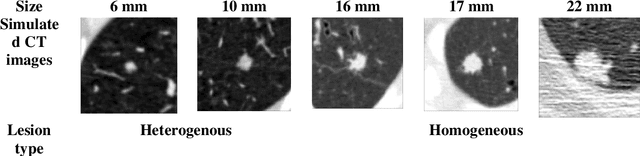
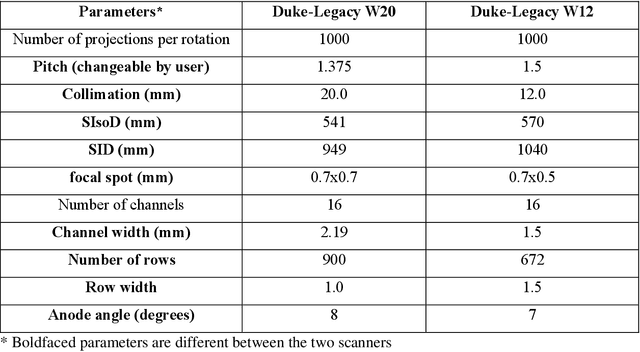
Abstract:Importance: The efficacy of lung cancer screening can be significantly impacted by the imaging modality used. This Virtual Lung Screening Trial (VLST) addresses the critical need for precision in lung cancer diagnostics and the potential for reducing unnecessary radiation exposure in clinical settings. Objectives: To establish a virtual imaging trial (VIT) platform that accurately simulates real-world lung screening trials (LSTs) to assess the diagnostic accuracy of CT and CXR modalities. Design, Setting, and Participants: Utilizing computational models and machine learning algorithms, we created a diverse virtual patient population. The cohort, designed to mirror real-world demographics, was assessed using virtual imaging techniques that reflect historical imaging technologies. Main Outcomes and Measures: The primary outcome was the difference in the Area Under the Curve (AUC) for CT and CXR modalities across lesion types and sizes. Results: The study analyzed 298 CT and 313 CXR simulated images from 313 virtual patients, with a lesion-level AUC of 0.81 (95% CI: 0.78-0.84) for CT and 0.55 (95% CI: 0.53-0.56) for CXR. At the patient level, CT demonstrated an AUC of 0.85 (95% CI: 0.80-0.89), compared to 0.53 (95% CI: 0.47-0.60) for CXR. Subgroup analyses indicated CT's superior performance in detecting homogeneous lesions (AUC of 0.97 for lesion-level) and heterogeneous lesions (AUC of 0.71 for lesion-level) as well as in identifying larger nodules (AUC of 0.98 for nodules > 8 mm). Conclusion and Relevance: The VIT platform validated the superior diagnostic accuracy of CT over CXR, especially for smaller nodules, underscoring its potential to replicate real clinical imaging trials. These findings advocate for the integration of virtual trials in the evaluation and improvement of imaging-based diagnostic tools.
Large Intestine 3D Shape Refinement Using Point Diffusion Models for Digital Phantom Generation
Sep 15, 2023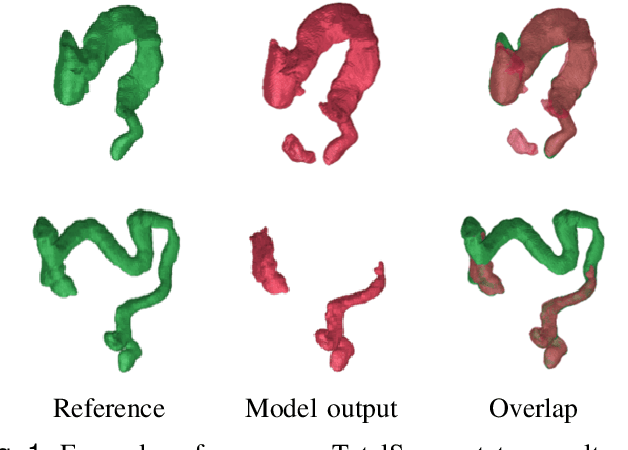
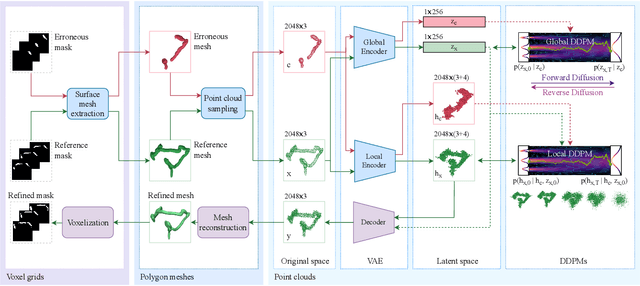
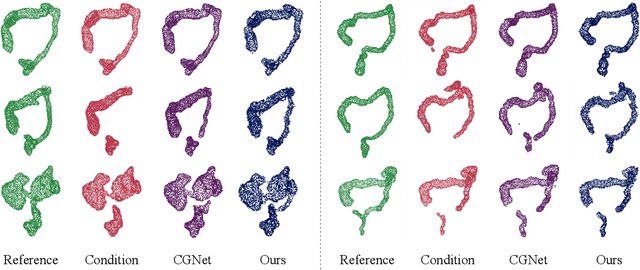
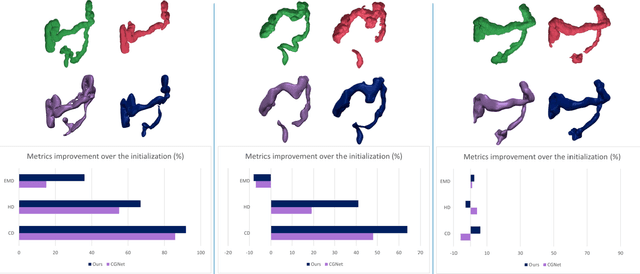
Abstract:Accurate 3D modeling of human organs plays a crucial role in building computational phantoms for virtual imaging trials. However, generating anatomically plausible reconstructions of organ surfaces from computed tomography scans remains challenging for many structures in the human body. This challenge is particularly evident when dealing with the large intestine. In this study, we leverage recent advancements in geometric deep learning and denoising diffusion probabilistic models to refine the segmentation results of the large intestine. We begin by representing the organ as point clouds sampled from the surface of the 3D segmentation mask. Subsequently, we employ a hierarchical variational autoencoder to obtain global and local latent representations of the organ's shape. We train two conditional denoising diffusion models in the hierarchical latent space to perform shape refinement. To further enhance our method, we incorporate a state-of-the-art surface reconstruction model, allowing us to generate smooth meshes from the obtained complete point clouds. Experimental results demonstrate the effectiveness of our approach in capturing both the global distribution of the organ's shape and its fine details. Our complete refinement pipeline demonstrates remarkable enhancements in surface representation compared to the initial segmentation, reducing the Chamfer distance by 70%, the Hausdorff distance by 32%, and the Earth Mover's distance by 6%. By combining geometric deep learning, denoising diffusion models, and advanced surface reconstruction techniques, our proposed method offers a promising solution for accurately modeling the large intestine's surface and can easily be extended to other anatomical structures.
Data diversity and virtual imaging in AI-based diagnosis: A case study based on COVID-19
Aug 17, 2023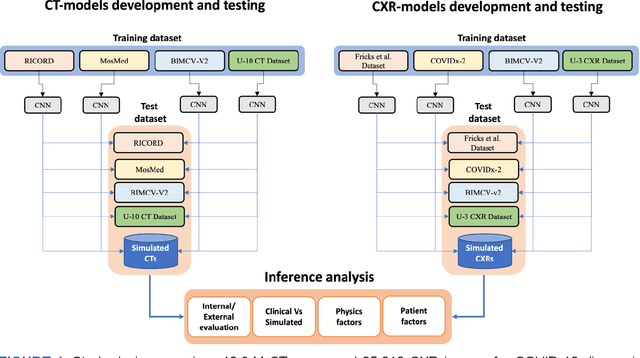
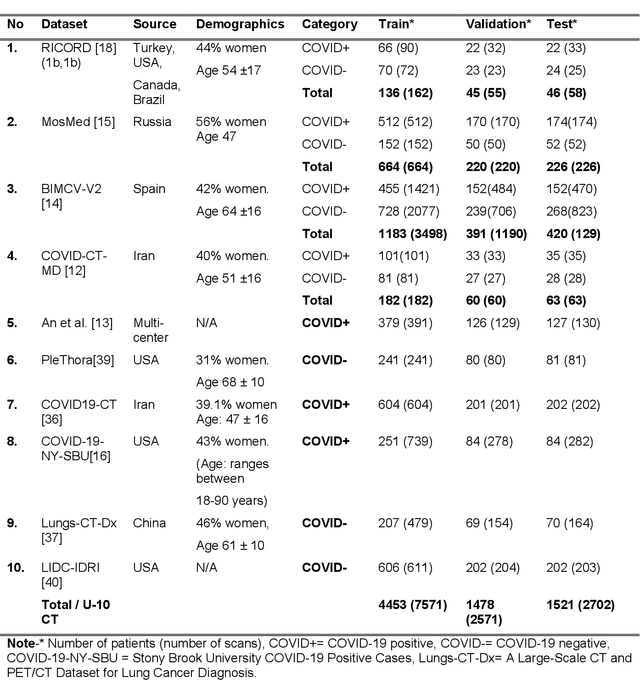
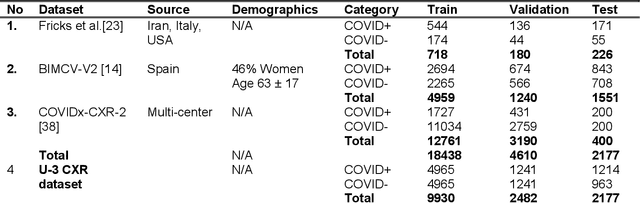
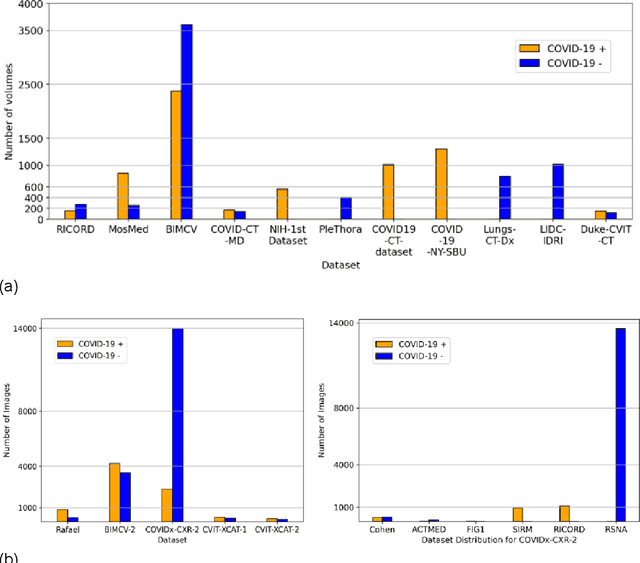
Abstract:Many studies have investigated deep-learning-based artificial intelligence (AI) models for medical imaging diagnosis of the novel coronavirus (COVID-19), with many reports of near-perfect performance. However, variability in performance and underlying data biases raise concerns about clinical generalizability. This retrospective study involved the development and evaluation of artificial intelligence (AI) models for COVID-19 diagnosis using both diverse clinical and virtually generated medical images. In addition, we conducted a virtual imaging trial to assess how AI performance is affected by several patient- and physics-based factors, including the extent of disease, radiation dose, and imaging modality of computed tomography (CT) and chest radiography (CXR). AI performance was strongly influenced by dataset characteristics including quantity, diversity, and prevalence, leading to poor generalization with up to 20% drop in receiver operating characteristic area under the curve. Model performance on virtual CT and CXR images was comparable to overall results on clinical data. Imaging dose proved to have negligible influence on the results, but the extent of the disease had a marked affect. CT results were consistently superior to those from CXR. Overall, the study highlighted the significant impact of dataset characteristics and disease extent on COVID assessment, and the relevance and potential role of virtual imaging trial techniques on developing effective evaluation of AI algorithms and facilitating translation into diagnostic practice.
Virtual vs. Reality: External Validation of COVID-19 Classifiers using XCAT Phantoms for Chest Computed Tomography
Mar 07, 2022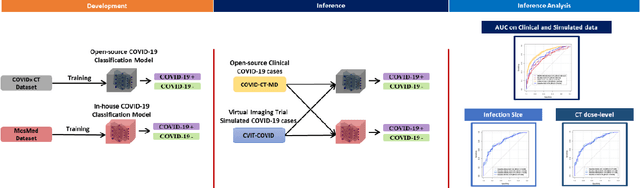
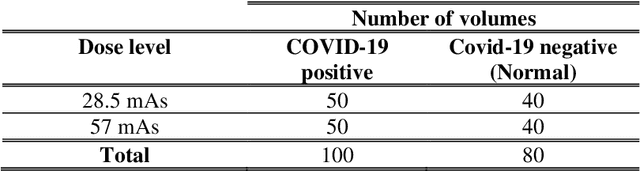
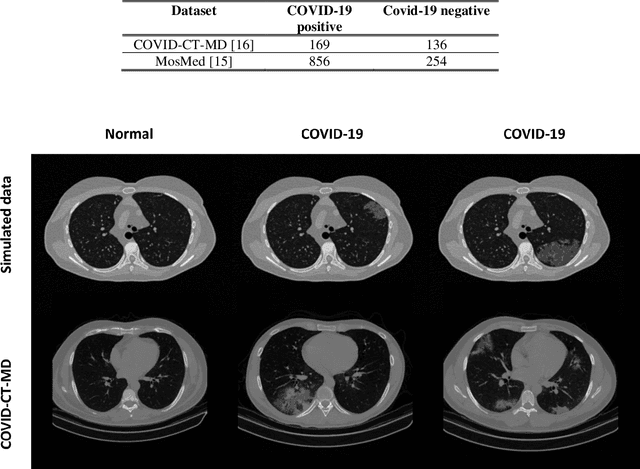
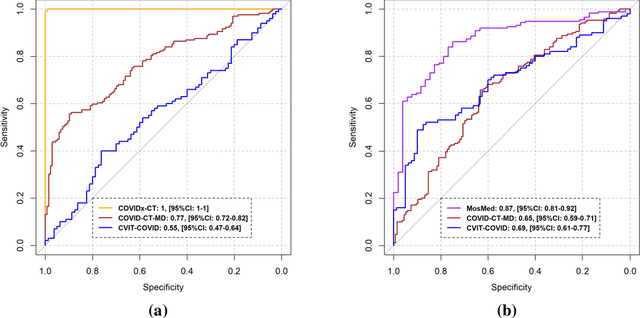
Abstract:Research studies of artificial intelligence models in medical imaging have been hampered by poor generalization. This problem has been especially concerning over the last year with numerous applications of deep learning for COVID-19 diagnosis. Virtual imaging trials (VITs) could provide a solution for objective evaluation of these models. In this work utilizing the VITs, we created the CVIT-COVID dataset including 180 virtually imaged computed tomography (CT) images from simulated COVID-19 and normal phantom models under different COVID-19 morphology and imaging properties. We evaluated the performance of an open-source, deep-learning model from the University of Waterloo trained with multi-institutional data and an in-house model trained with the open clinical dataset called MosMed. We further validated the model's performance against open clinical data of 305 CT images to understand virtual vs. real clinical data performance. The open-source model was published with nearly perfect performance on the original Waterloo dataset but showed a consistent performance drop in external testing on another clinical dataset (AUC=0.77) and our simulated CVIT-COVID dataset (AUC=0.55). The in-house model achieved an AUC of 0.87 while testing on the internal test set (MosMed test set). However, performance dropped to an AUC of 0.65 and 0.69 when evaluated on clinical and our simulated CVIT-COVID dataset. The VIT framework offered control over imaging conditions, allowing us to show there was no change in performance as CT exposure was changed from 28.5 to 57 mAs. The VIT framework also provided voxel-level ground truth, revealing that performance of in-house model was much higher at AUC=0.87 for diffuse COVID-19 infection size >2.65% lung volume versus AUC=0.52 for focal disease with <2.65% volume. The virtual imaging framework enabled these uniquely rigorous analyses of model performance.
Quality or Quantity: Toward a Unified Approach for Multi-organ Segmentation in Body CT
Mar 03, 2022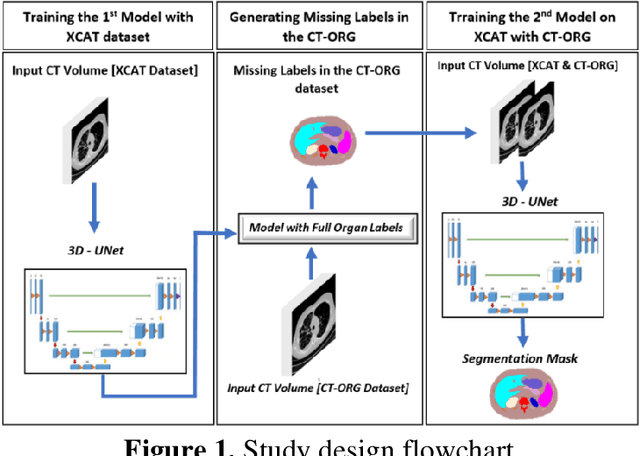
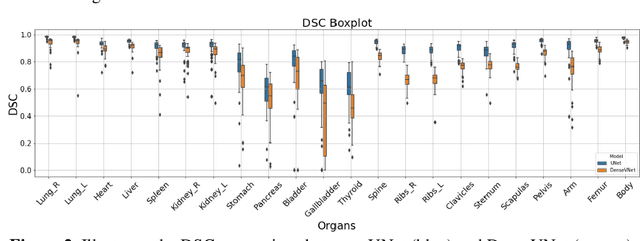
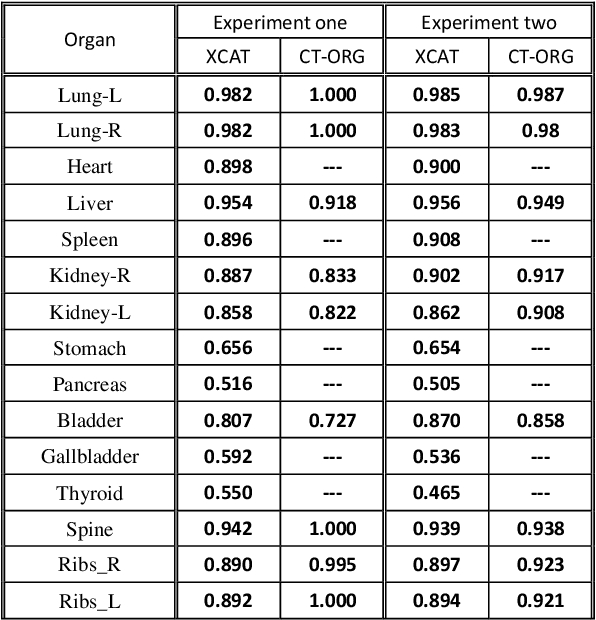
Abstract:Organ segmentation of medical images is a key step in virtual imaging trials. However, organ segmentation datasets are limited in terms of quality (because labels cover only a few organs) and quantity (since case numbers are limited). In this study, we explored the tradeoffs between quality and quantity. Our goal is to create a unified approach for multi-organ segmentation of body CT, which will facilitate the creation of large numbers of accurate virtual phantoms. Initially, we compared two segmentation architectures, 3D-Unet and DenseVNet, which were trained using XCAT data that is fully labeled with 22 organs, and chose the 3D-Unet as the better performing model. We used the XCAT-trained model to generate pseudo-labels for the CT-ORG dataset that has only 7 organs segmented. We performed two experiments: First, we trained 3D-UNet model on the XCAT dataset, representing quality data, and tested it on both XCAT and CT-ORG datasets. Second, we trained 3D-UNet after including the CT-ORG dataset into the training set to have more quantity. Performance improved for segmentation in the organs where we have true labels in both datasets and degraded when relying on pseudo-labels. When organs were labeled in both datasets, Exp-2 improved Average DSC in XCAT and CT-ORG by 1. This demonstrates that quality data is the key to improving the model's performance.
Co-occurring Diseases Heavily Influence the Performance of Weakly Supervised Learning Models for Classification of Chest CT
Feb 23, 2022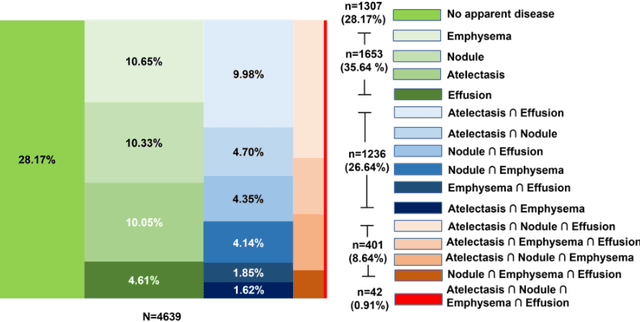
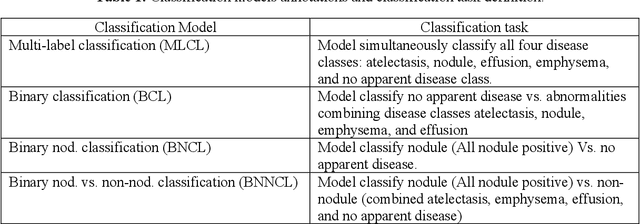
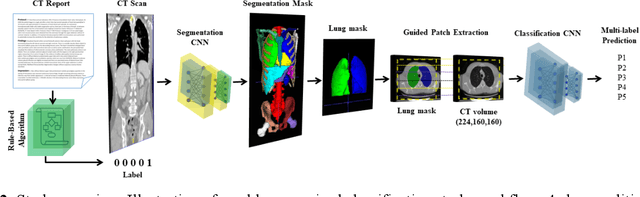
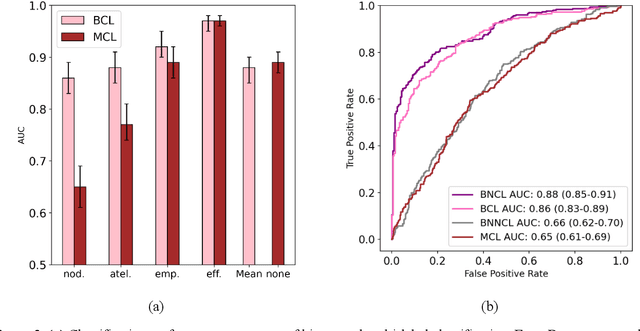
Abstract:Despite the potential of weakly supervised learning to automatically annotate massive amounts of data, little is known about its limitations for use in computer-aided diagnosis (CAD). For CT specifically, interpreting the performance of CAD algorithms can be challenging given the large number of co-occurring diseases. This paper examines the effect of co-occurring diseases when training classification models by weakly supervised learning, specifically by comparing multi-label and multiple binary classifiers using the same training data. Our results demonstrated that the binary model outperformed the multi-label classification in every disease category in terms of AUC. However, this performance was heavily influenced by co-occurring diseases in the binary model, suggesting it did not always learn the correct appearance of the specific disease. For example, binary classification of lung nodules resulted in an AUC of < 0.65 when there were no other co-occurring diseases, but when lung nodules co-occurred with emphysema, the performance reached AUC > 0.80. We hope this paper revealed the complexity of interpreting disease classification performance in weakly supervised models and will encourage researchers to examine the effect of co-occurring diseases on classification performance in the future.
iPhantom: a framework for automated creation of individualized computational phantoms and its application to CT organ dosimetry
Aug 20, 2020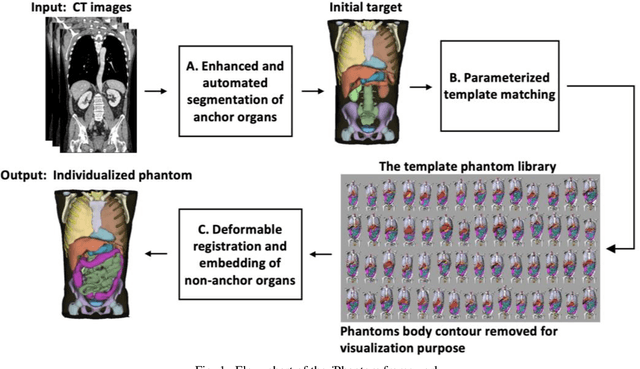
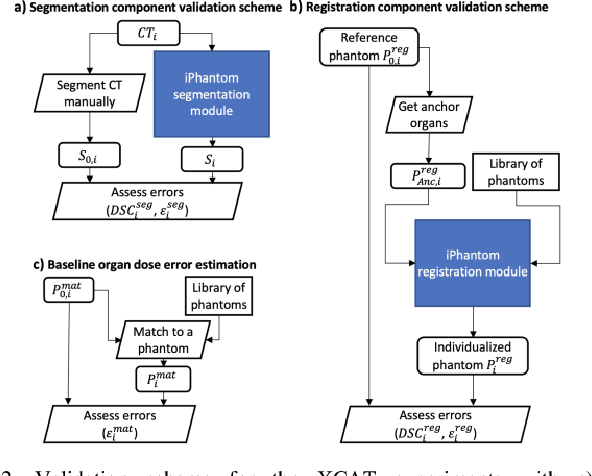
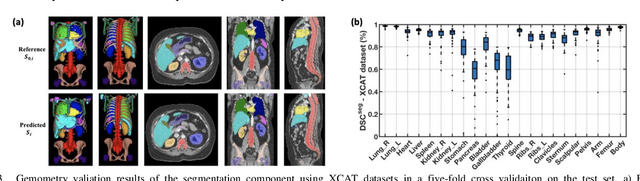
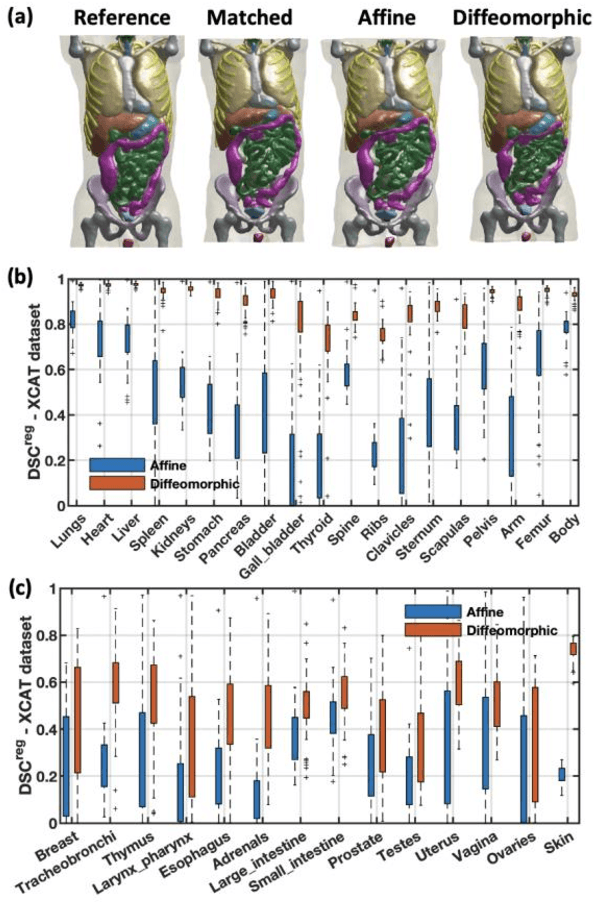
Abstract:Objective: This study aims to develop and validate a novel framework, iPhantom, for automated creation of patient-specific phantoms or digital-twins (DT) using patient medical images. The framework is applied to assess radiation dose to radiosensitive organs in CT imaging of individual patients. Method: From patient CT images, iPhantom segments selected anchor organs (e.g. liver, bones, pancreas) using a learning-based model developed for multi-organ CT segmentation. Organs challenging to segment (e.g. intestines) are incorporated from a matched phantom template, using a diffeomorphic registration model developed for multi-organ phantom-voxels. The resulting full-patient phantoms are used to assess organ doses during routine CT exams. Result: iPhantom was validated on both the XCAT (n=50) and an independent clinical (n=10) dataset with similar accuracy. iPhantom precisely predicted all organ locations with good accuracy of Dice Similarity Coefficients (DSC) >0.6 for anchor organs and DSC of 0.3-0.9 for all other organs. iPhantom showed less than 10% dose errors for the majority of organs, which was notably superior to the state-of-the-art baseline method (20-35% dose errors). Conclusion: iPhantom enables automated and accurate creation of patient-specific phantoms and, for the first time, provides sufficient and automated patient-specific dose estimates for CT dosimetry. Significance: The new framework brings the creation and application of CHPs to the level of individual CHPs through automation, achieving a wider and precise organ localization, paving the way for clinical monitoring, and personalized optimization, and large-scale research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge