Rafael B. Fricks
Virtual vs. Reality: External Validation of COVID-19 Classifiers using XCAT Phantoms for Chest Computed Tomography
Mar 07, 2022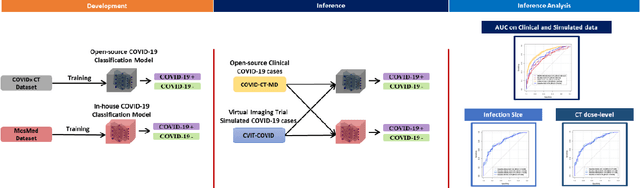
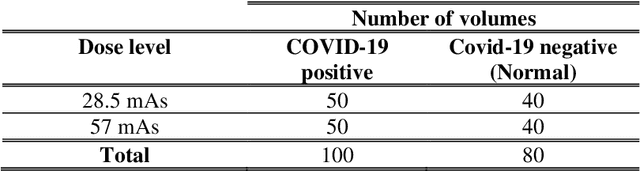
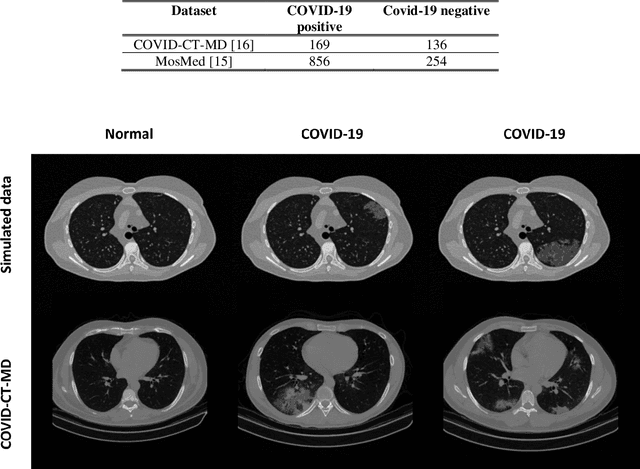
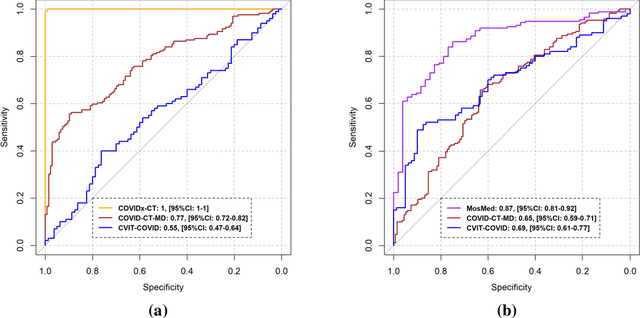
Abstract:Research studies of artificial intelligence models in medical imaging have been hampered by poor generalization. This problem has been especially concerning over the last year with numerous applications of deep learning for COVID-19 diagnosis. Virtual imaging trials (VITs) could provide a solution for objective evaluation of these models. In this work utilizing the VITs, we created the CVIT-COVID dataset including 180 virtually imaged computed tomography (CT) images from simulated COVID-19 and normal phantom models under different COVID-19 morphology and imaging properties. We evaluated the performance of an open-source, deep-learning model from the University of Waterloo trained with multi-institutional data and an in-house model trained with the open clinical dataset called MosMed. We further validated the model's performance against open clinical data of 305 CT images to understand virtual vs. real clinical data performance. The open-source model was published with nearly perfect performance on the original Waterloo dataset but showed a consistent performance drop in external testing on another clinical dataset (AUC=0.77) and our simulated CVIT-COVID dataset (AUC=0.55). The in-house model achieved an AUC of 0.87 while testing on the internal test set (MosMed test set). However, performance dropped to an AUC of 0.65 and 0.69 when evaluated on clinical and our simulated CVIT-COVID dataset. The VIT framework offered control over imaging conditions, allowing us to show there was no change in performance as CT exposure was changed from 28.5 to 57 mAs. The VIT framework also provided voxel-level ground truth, revealing that performance of in-house model was much higher at AUC=0.87 for diffuse COVID-19 infection size >2.65% lung volume versus AUC=0.52 for focal disease with <2.65% volume. The virtual imaging framework enabled these uniquely rigorous analyses of model performance.
Automatic phantom test pattern classification through transfer learning with deep neural networks
Jan 22, 2020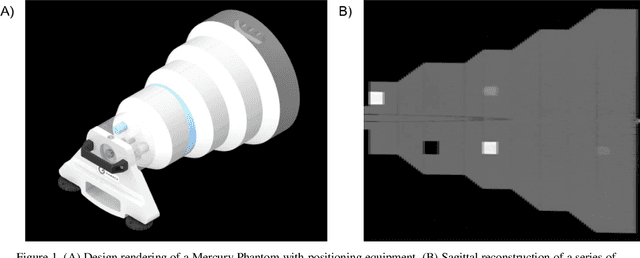
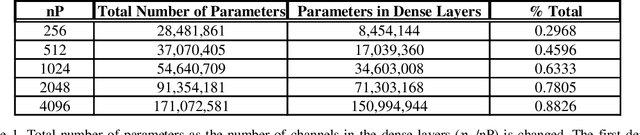
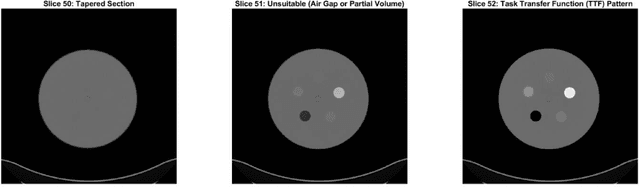
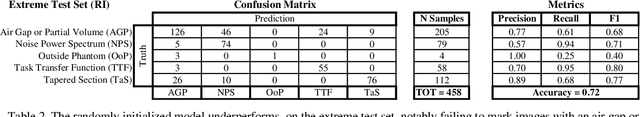
Abstract:Imaging phantoms are test patterns used to measure image quality in computer tomography (CT) systems. A new phantom platform (Mercury Phantom, Gammex) provides test patterns for estimating the task transfer function (TTF) or noise power spectrum (NPF) and simulates different patient sizes. Determining which image slices are suitable for analysis currently requires manual annotation of these patterns by an expert, as subtle defects may make an image unsuitable for measurement. We propose a method of automatically classifying these test patterns in a series of phantom images using deep learning techniques. By adapting a convolutional neural network based on the VGG19 architecture with weights trained on ImageNet, we use transfer learning to produce a classifier for this domain. The classifier is trained and evaluated with over 3,500 phantom images acquired at a university medical center. Input channels for color images are successfully adapted to convey contextual information for phantom images. A series of ablation studies are employed to verify design aspects of the classifier and evaluate its performance under varying training conditions. Our solution makes extensive use of image augmentation to produce a classifier that accurately classifies typical phantom images with 98% accuracy, while maintaining as much as 86% accuracy when the phantom is improperly imaged.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge