Dhrubajyoti Ghosh
Integrating Causal Inference with Graph Neural Networks for Alzheimer's Disease Analysis
Nov 18, 2025Abstract:Deep graph learning has advanced Alzheimer's (AD) disease classification from MRI, but most models remain correlational, confounding demographic and genetic factors with disease specific features. We present Causal-GCN, an interventional graph convolutional framework that integrates do-calculus-based back-door adjustment to identify brain regions exerting stable causal influence on AD progression. Each subject's MRI is represented as a structural connectome where nodes denote cortical and subcortical regions and edges encode anatomical connectivity. Confounders such as age, sec, and APOE4 genotype are summarized via principal components and included in the causal adjustment set. After training, interventions on individual regions are simulated by serving their incoming edges and altering node features to estimate average causal effects on disease probability. Applied to 484 subjects from the ADNI cohort, Causal-GCN achieves performance comparable to baseline GNNs while providing interpretable causal effect rankings that highlight posterior, cingulate, and insular hubs consistent with established AD neuropathology.
Ensemble Survival Analysis for Preclinical Cognitive Decline Prediction in Alzheimer's Disease Using Longitudinal Biomarkers
Mar 20, 2025Abstract:Predicting the risk of clinical progression from cognitively normal (CN) status to mild cognitive impairment (MCI) or Alzheimer's disease (AD) is critical for early intervention in Alzheimer's disease (AD). Traditional survival models often fail to capture complex longitudinal biomarker patterns associated with disease progression. We propose an ensemble survival analysis framework integrating multiple survival models to improve early prediction of clinical progression in initially cognitively normal individuals. We analyzed longitudinal biomarker data from the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort, including 721 participants, limiting analysis to up to three visits (baseline, 6-month follow-up, 12-month follow-up). Of these, 142 (19.7%) experienced clinical progression to MCI or AD. Our approach combined penalized Cox regression (LASSO, Elastic Net) with advanced survival models (Random Survival Forest, DeepSurv, XGBoost). Model predictions were aggregated using ensemble averaging and Bayesian Model Averaging (BMA). Predictive performance was assessed using Harrell's concordance index (C-index) and time-dependent area under the curve (AUC). The ensemble model achieved a peak C-index of 0.907 and an integrated time-dependent AUC of 0.904, outperforming baseline-only models (C-index 0.608). One follow-up visit after baseline significantly improved prediction accuracy (48.1% C-index, 48.2% AUC gains), while adding a second follow-up provided only marginal gains (2.1% C-index, 2.7% AUC). Our ensemble survival framework effectively integrates diverse survival models and aggregation techniques to enhance early prediction of preclinical AD progression. These findings highlight the importance of leveraging longitudinal biomarker data, particularly one follow-up visit, for accurate risk stratification and personalized intervention strategies.
XCAT-2.0: A Comprehensive Library of Personalized Digital Twins Derived from CT Scans
May 18, 2024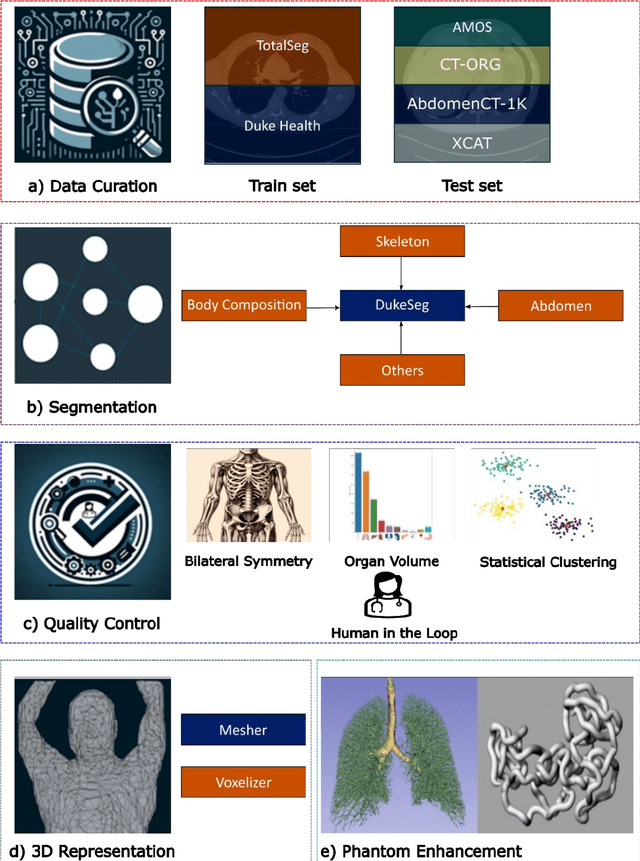
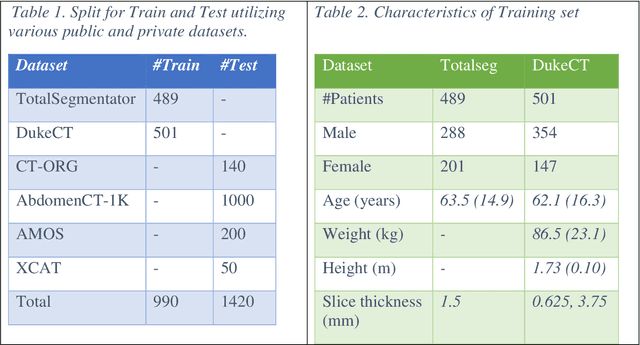
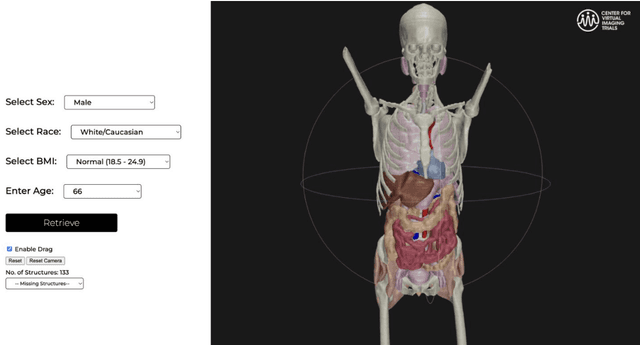
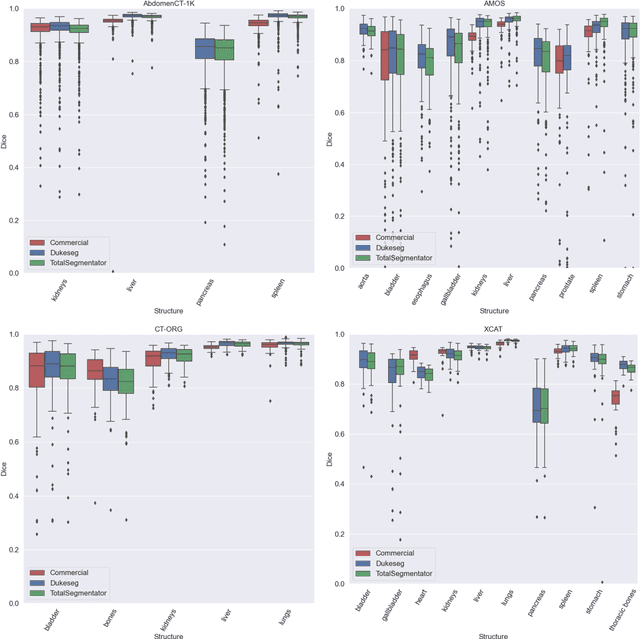
Abstract:Virtual Imaging Trials (VIT) offer a cost-effective and scalable approach for evaluating medical imaging technologies. Computational phantoms, which mimic real patient anatomy and physiology, play a central role in VIT. However, the current libraries of computational phantoms face limitations, particularly in terms of sample size and diversity. Insufficient representation of the population hampers accurate assessment of imaging technologies across different patient groups. Traditionally, phantoms were created by manual segmentation, which is a laborious and time-consuming task, impeding the expansion of phantom libraries. This study presents a framework for realistic computational phantom modeling using a suite of four deep learning segmentation models, followed by three forms of automated organ segmentation quality control. Over 2500 computational phantoms with up to 140 structures illustrating a sophisticated approach to detailed anatomical modeling are released. Phantoms are available in both voxelized and surface mesh formats. The framework is aggregated with an in-house CT scanner simulator to produce realistic CT images. The framework can potentially advance virtual imaging trials, facilitating comprehensive and reliable evaluations of medical imaging technologies. Phantoms may be requested at https://cvit.duke.edu/resources/, code, model weights, and sample CT images are available at https://xcat-2.github.io.
VLST: Virtual Lung Screening Trial for Lung Cancer Detection Using Virtual Imaging Trial
Apr 17, 2024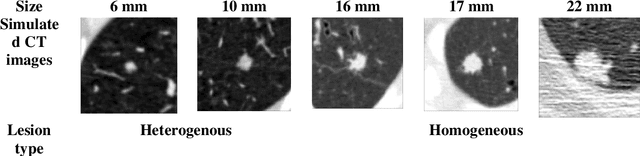
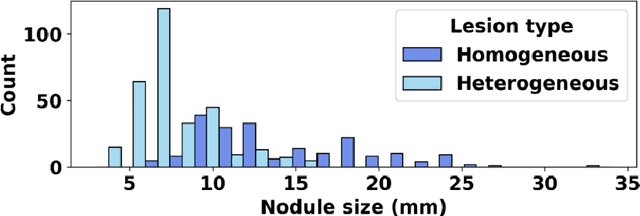
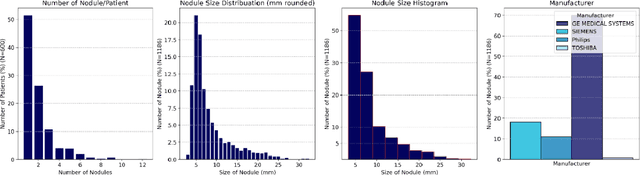
Abstract:Importance: The efficacy of lung cancer screening can be significantly impacted by the imaging modality used. This Virtual Lung Screening Trial (VLST) addresses the critical need for precision in lung cancer diagnostics and the potential for reducing unnecessary radiation exposure in clinical settings. Objectives: To establish a virtual imaging trial (VIT) platform that accurately simulates real-world lung screening trials (LSTs) to assess the diagnostic accuracy of CT and CXR modalities. Design, Setting, and Participants: Utilizing computational models and machine learning algorithms, we created a diverse virtual patient population. The cohort, designed to mirror real-world demographics, was assessed using virtual imaging techniques that reflect historical imaging technologies. Main Outcomes and Measures: The primary outcome was the difference in the Area Under the Curve (AUC) for CT and CXR modalities across lesion types and sizes. Results: The study analyzed 298 CT and 313 CXR simulated images from 313 virtual patients, with a lesion-level AUC of 0.81 (95% CI: 0.78-0.84) for CT and 0.55 (95% CI: 0.53-0.56) for CXR. At the patient level, CT demonstrated an AUC of 0.85 (95% CI: 0.80-0.89), compared to 0.53 (95% CI: 0.47-0.60) for CXR. Subgroup analyses indicated CT's superior performance in detecting homogeneous lesions (AUC of 0.97 for lesion-level) and heterogeneous lesions (AUC of 0.71 for lesion-level) as well as in identifying larger nodules (AUC of 0.98 for nodules > 8 mm). Conclusion and Relevance: The VIT platform validated the superior diagnostic accuracy of CT over CXR, especially for smaller nodules, underscoring its potential to replicate real clinical imaging trials. These findings advocate for the integration of virtual trials in the evaluation and improvement of imaging-based diagnostic tools.
Prism: Private Verifiable Set Computation over Multi-Owner Outsourced Databases
Apr 07, 2021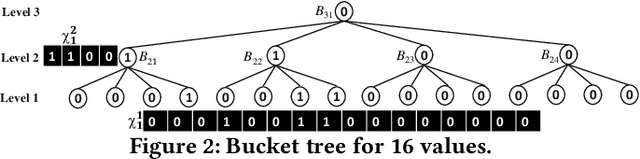
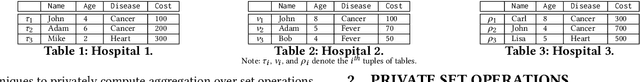
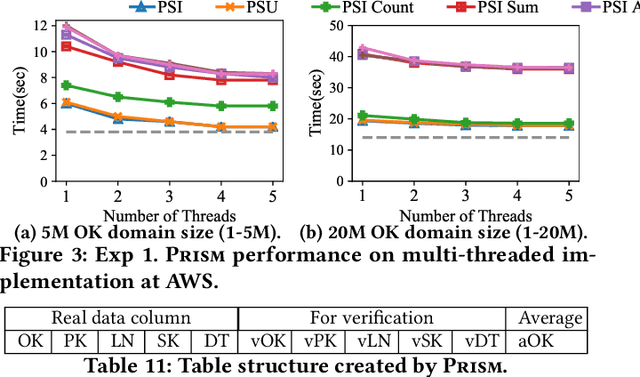
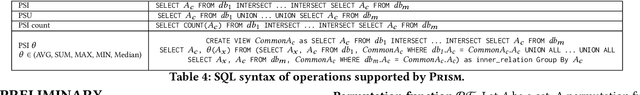
Abstract:This paper proposes Prism, a secret sharing based approach to compute private set operations (i.e., intersection and union), as well as aggregates over outsourced databases belonging to multiple owners. Prism enables data owners to pre-load the data onto non-colluding servers and exploits the additive and multiplicative properties of secret-shares to compute the above-listed operations in (at most) two rounds of communication between the servers (storing the secret-shares) and the querier, resulting in a very efficient implementation. Also, Prism does not require communication among the servers and supports result verification techniques for each operation to detect malicious adversaries. Experimental results show that Prism scales both in terms of the number of data owners and database sizes, to which prior approaches do not scale.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge