Geoffrey D. Rubin
Organ-Aware Attention Improves CT Triage and Classification
Jan 19, 2026Abstract:There is an urgent need for triage and classification of high-volume medical imaging modalities such as computed tomography (CT), which can improve patient care and mitigate radiologist burnout. Study-level CT triage requires calibrated predictions with localized evidence; however, off-the-shelf Vision Language Models (VLM) struggle with 3D anatomy, protocol shifts, and noisy report supervision. This study used the two largest publicly available chest CT datasets: CT-RATE and RADCHEST-CT (held-out external test set). Our carefully tuned supervised baseline (instantiated as a simple Global Average Pooling head) establishes a new supervised state of the art, surpassing all reported linear-probe VLMs. Building on this baseline, we present ORACLE-CT, an encoder-agnostic, organ-aware head that pairs Organ-Masked Attention (mask-restricted, per-organ pooling that yields spatial evidence) with Organ-Scalar Fusion (lightweight fusion of normalized volume and mean-HU cues). In the chest setting, ORACLE-CT masked attention model achieves AUROC 0.86 on CT-RATE; in the abdomen setting, on MERLIN (30 findings), our supervised baseline exceeds a reproduced zero-shot VLM baseline obtained by running publicly released weights through our pipeline, and adding masked attention plus scalar fusion further improves performance to AUROC 0.85. Together, these results deliver state-of-the-art supervised classification performance across both chest and abdomen CT under a unified evaluation protocol. The source code is available at https://github.com/lavsendahal/oracle-ct.
What limits performance of weakly supervised deep learning for chest CT classification?
Feb 06, 2024Abstract:Weakly supervised learning with noisy data has drawn attention in the medical imaging community due to the sparsity of high-quality disease labels. However, little is known about the limitations of such weakly supervised learning and the effect of these constraints on disease classification performance. In this paper, we test the effects of such weak supervision by examining model tolerance for three conditions. First, we examined model tolerance for noisy data by incrementally increasing error in the labels within the training data. Second, we assessed the impact of dataset size by varying the amount of training data. Third, we compared performance differences between binary and multi-label classification. Results demonstrated that the model could endure up to 10% added label error before experiencing a decline in disease classification performance. Disease classification performance steadily rose as the amount of training data was increased for all disease classes, before experiencing a plateau in performance at 75% of training data. Last, the binary model outperformed the multilabel model in every disease category. However, such interpretations may be misleading, as the binary model was heavily influenced by co-occurring diseases and may not have learned the specific features of the disease in the image. In conclusion, this study may help the medical imaging community understand the benefits and risks of weak supervision with noisy labels. Such studies demonstrate the need to build diverse, large-scale datasets and to develop explainable and responsible AI.
Co-occurring Diseases Heavily Influence the Performance of Weakly Supervised Learning Models for Classification of Chest CT
Feb 23, 2022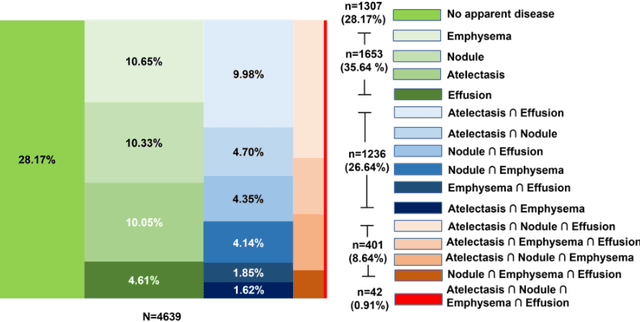
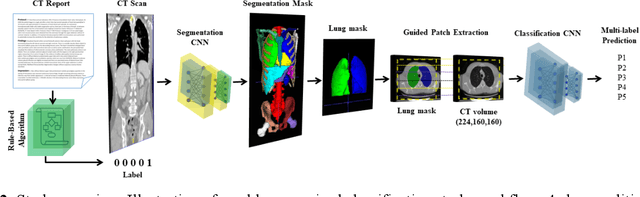
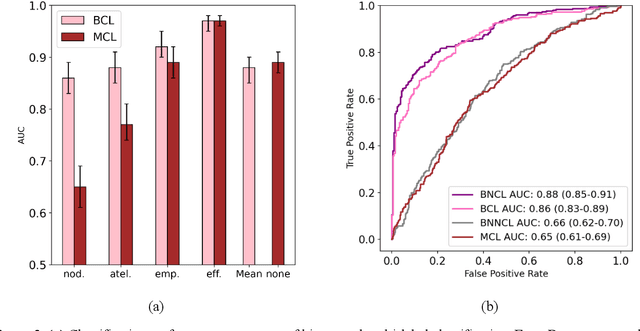
Abstract:Despite the potential of weakly supervised learning to automatically annotate massive amounts of data, little is known about its limitations for use in computer-aided diagnosis (CAD). For CT specifically, interpreting the performance of CAD algorithms can be challenging given the large number of co-occurring diseases. This paper examines the effect of co-occurring diseases when training classification models by weakly supervised learning, specifically by comparing multi-label and multiple binary classifiers using the same training data. Our results demonstrated that the binary model outperformed the multi-label classification in every disease category in terms of AUC. However, this performance was heavily influenced by co-occurring diseases in the binary model, suggesting it did not always learn the correct appearance of the specific disease. For example, binary classification of lung nodules resulted in an AUC of < 0.65 when there were no other co-occurring diseases, but when lung nodules co-occurred with emphysema, the performance reached AUC > 0.80. We hope this paper revealed the complexity of interpreting disease classification performance in weakly supervised models and will encourage researchers to examine the effect of co-occurring diseases on classification performance in the future.
Multi-Label Annotation of Chest Abdomen Pelvis Computed Tomography Text Reports Using Deep Learning
Feb 25, 2021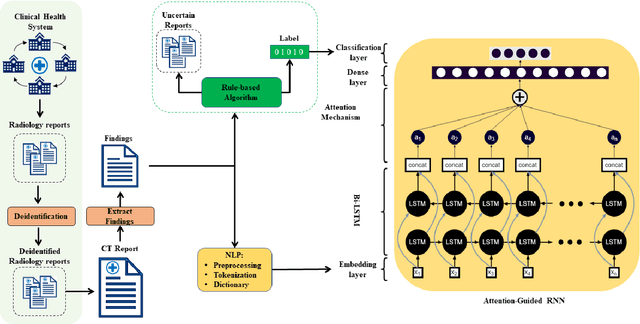
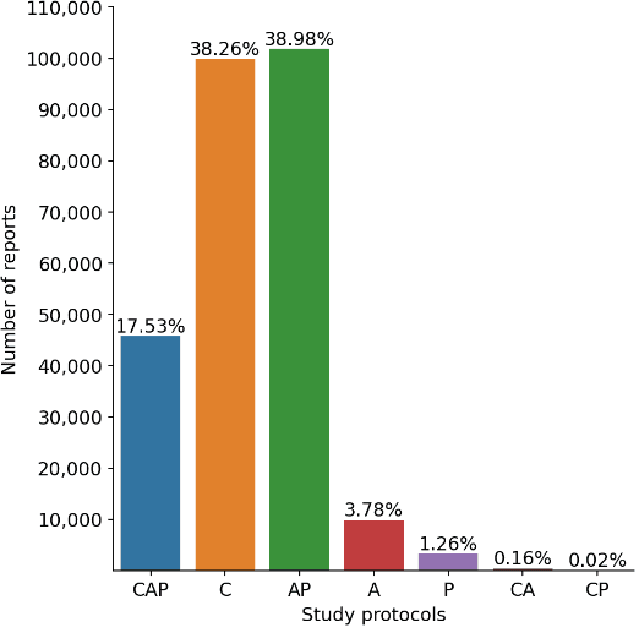
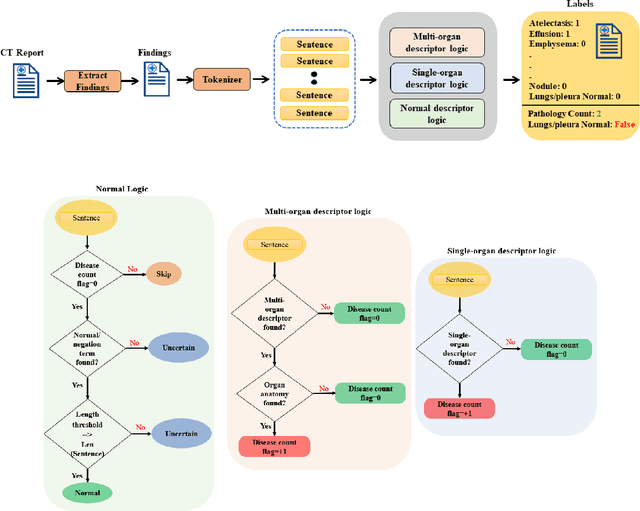
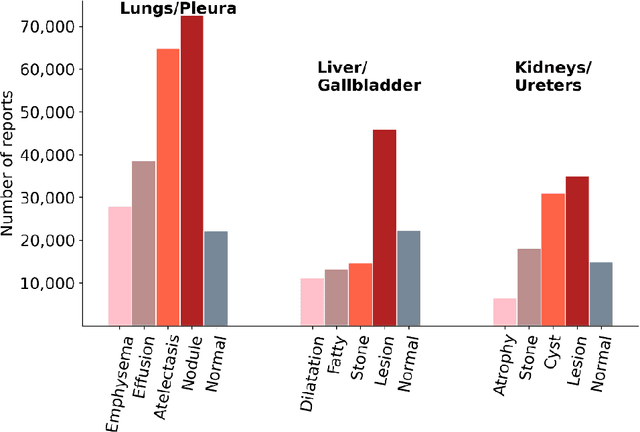
Abstract:To develop a high throughput multi-label annotator for body Computed Tomography (CT) reports that can be applied to a variety of diseases, organs, and cases. First, we used a dictionary approach to develop a rule-based algorithm (RBA) for extraction of disease labels from radiology text reports. We targeted three organ systems (lungs/pleura, liver/gallbladder, kidneys/ureters) with four diseases per system based on their prevalence in our dataset. To expand the algorithm beyond pre-defined keywords, an attention-guided recurrent neural network (RNN) was trained using the RBA-extracted labels to classify the reports as being positive for one or more diseases or normal for each organ system. Confounding effects on model performance were evaluated using random or pre-trained embedding as well as different sizes of training datasets. Performance was evaluated using the receiver operating characteristic (ROC) area under the curve (AUC) against 2,158 manually obtained labels. Our model extracted disease labels from 261,229 radiology reports of 112,501 unique subjects. Pre-trained models outperformed random embedding across all diseases. As the training dataset size was reduced, performance was robust except for a few diseases with relatively small number of cases. Pre-trained Classification AUCs achieved > 0.95 for all five disease outcomes across all three organ systems. Our label-extracting pipeline was able to encompass a variety of cases and diseases by generalizing beyond strict rules with exceptional accuracy. As a framework, this model can be easily adapted to enable automated labeling of hospital-scale medical data sets for training image-based disease classifiers.
Weakly Supervised 3D Classification of Chest CT using Aggregated Multi-Resolution Deep Segmentation Features
Oct 31, 2020Abstract:Weakly supervised disease classification of CT imaging suffers from poor localization owing to case-level annotations, where even a positive scan can hold hundreds to thousands of negative slices along multiple planes. Furthermore, although deep learning segmentation and classification models extract distinctly unique combinations of anatomical features from the same target class(es), they are typically seen as two independent processes in a computer-aided diagnosis (CAD) pipeline, with little to no feature reuse. In this research, we propose a medical classifier that leverages the semantic structural concepts learned via multi-resolution segmentation feature maps, to guide weakly supervised 3D classification of chest CT volumes. Additionally, a comparative analysis is drawn across two different types of feature aggregation to explore the vast possibilities surrounding feature fusion. Using a dataset of 1593 scans labeled on a case-level basis via rule-based model, we train a dual-stage convolutional neural network (CNN) to perform organ segmentation and binary classification of four representative diseases (emphysema, pneumonia/atelectasis, mass and nodules) in lungs. The baseline model, with separate stages for segmentation and classification, results in AUC of 0.791. Using identical hyperparameters, the connected architecture using static and dynamic feature aggregation improves performance to AUC of 0.832 and 0.851, respectively. This study advances the field in two key ways. First, case-level report data is used to weakly supervise a 3D CT classifier of multiple, simultaneous diseases for an organ. Second, segmentation and classification models are connected with two different feature aggregation strategies to enhance the classification performance.
Weakly Supervised Multi-Organ Multi-Disease Classification of Body CT Scans
Aug 03, 2020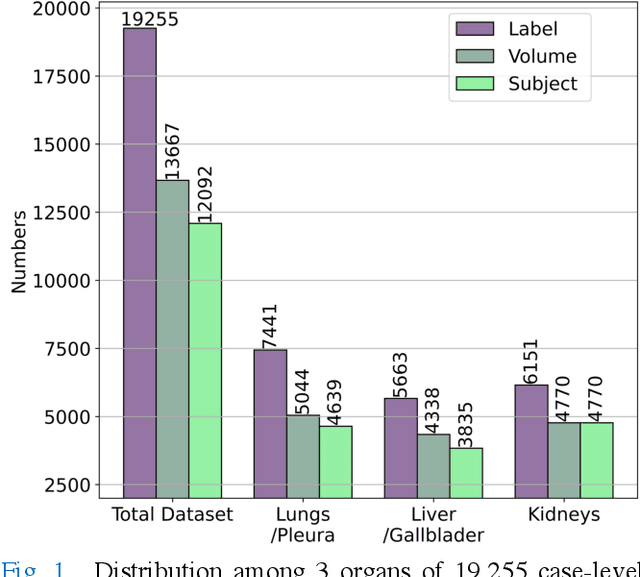
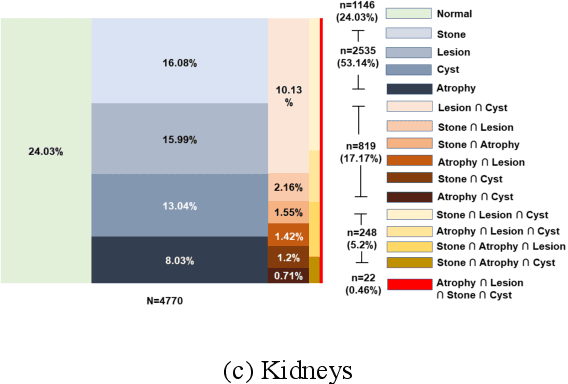
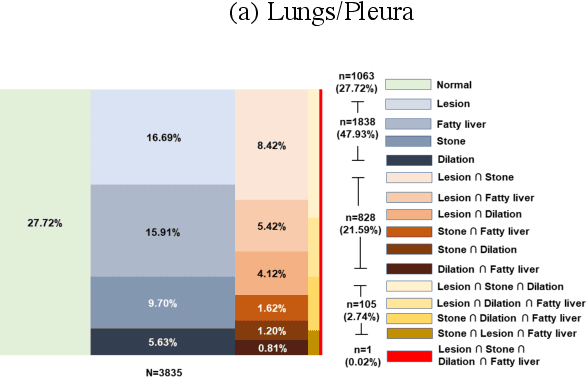
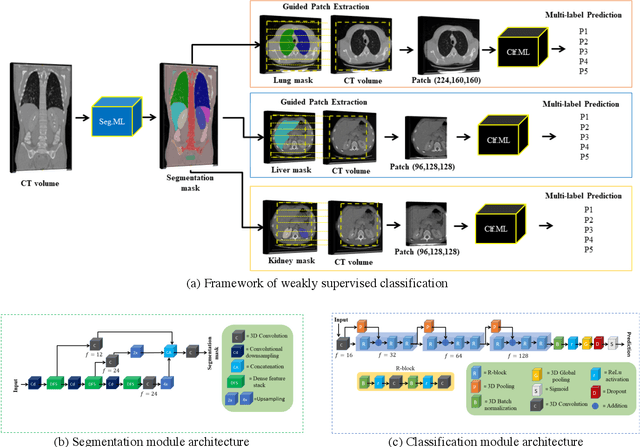
Abstract:We designed a multi-organ, multi-label disease classification algorithm for computed tomography (CT) scans using case-level labels from radiology text reports. A rule-based algorithm extracted 19,255 disease labels from reports of 13,667 body CT scans from 12,092 subjects. A 3D DenseVNet was trained to segment 3 organ systems: lungs/pleura, liver/gallbladder, and kidneys. From patches guided by segmentations, a 3D convolutional neural network provided multi-label disease classification for normality versus four common diseases per organ. The process was tested on 2,158 CT volumes with 2,875 manually obtained labels. Manual validation of the rulebased labels confirmed 91 to 99% accuracy. Results were characterized using the receiver operating characteristic area under the curve (AUC). Classification AUCs for lungs/pleura labels were as follows: atelectasis 0.77 (95% confidence intervals 0.74 to 0.81), nodule 0.65 (0.61 to 0.69), emphysema 0.89 (0.86 to 0.92), effusion 0.97 (0.96 to 0.98), and normal 0.89 (0.87 to 0.91). For liver/gallbladder, AUCs were: stone 0.62 (0.56 to 0.67), lesion 0.73 (0.69 to 0.77), dilation 0.87 (0.84 to 0.90), fatty 0.89 (0.86 to 0.92), and normal 0.82 (0.78 to 0.85). For kidneys, AUCs were: stone 0.83 (0.79 to 0.87), atrophy 0.92 (0.89 to 0.94), lesion 0.68 (0.64 to 0.72), cyst 0.70 (0.66 to 0.73), and normal 0.79 (0.75 to 0.83). In conclusion, by using automated extraction of disease labels from radiology reports, we created a weakly supervised, multi-organ, multi-disease classifier that can be easily adapted to efficiently leverage massive amounts of unannotated data associated with medical images.
Machine-Learning-Based Multiple Abnormality Prediction with Large-Scale Chest Computed Tomography Volumes
Feb 17, 2020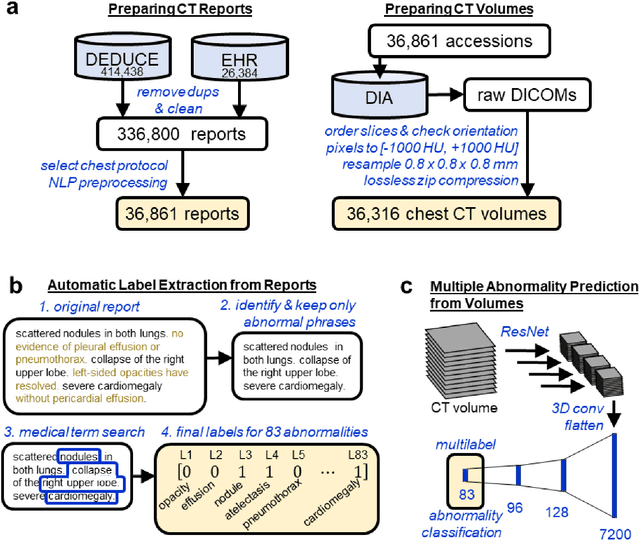

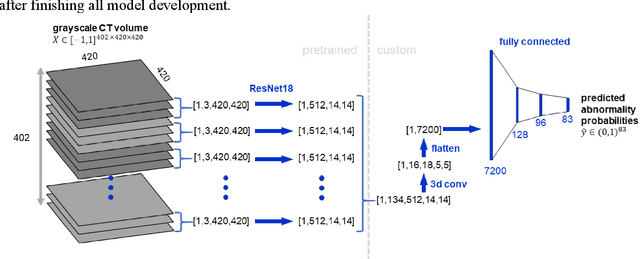
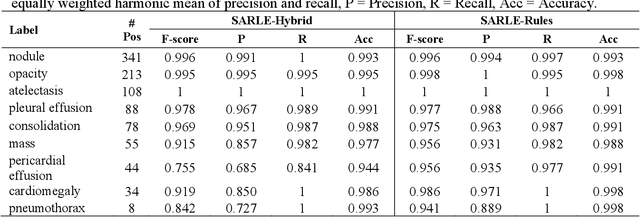
Abstract:Machine learning models for radiology benefit from large-scale data sets with high quality labels for abnormalities. We curated and analyzed a chest computed tomography (CT) data set of 36,316 volumes from 19,993 unique patients. This is the largest multiply-annotated volumetric medical imaging data set reported. To annotate this data set, we developed a rule-based method for automatically extracting abnormality labels from free-text radiology reports with an average F-score of 0.976 (min 0.941, max 1.0). We also developed a model for multi-organ, multi-disease classification of chest CT volumes that uses a deep convolutional neural network (CNN). This model reached a classification performance of AUROC greater than 0.90 for 18 abnormalities, with an average AUROC of 0.773 for all 83 abnormalities, demonstrating the feasibility of learning from unfiltered whole volume CT data. We show that training on more labels improves performance significantly: for a subset of 9 labels - nodule, opacity, atelectasis, pleural effusion, consolidation, mass, pericardial effusion, cardiomegaly, and pneumothorax - the model's average AUROC increased by 10% when the number of training labels was increased from 9 to all 83. All code for volume preprocessing, automated label extraction, and the volume abnormality prediction model will be made publicly available. The 36,316 CT volumes and labels will also be made publicly available pending institutional approval.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge