Multi-Label Annotation of Chest Abdomen Pelvis Computed Tomography Text Reports Using Deep Learning
Paper and Code
Feb 25, 2021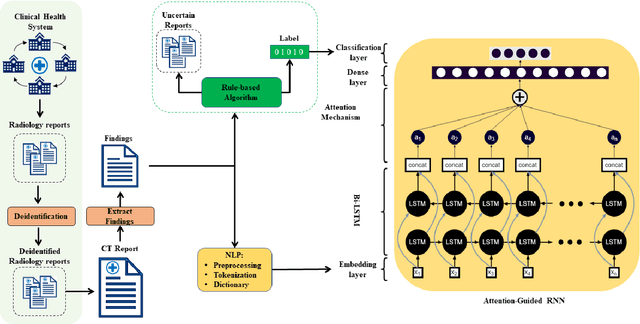
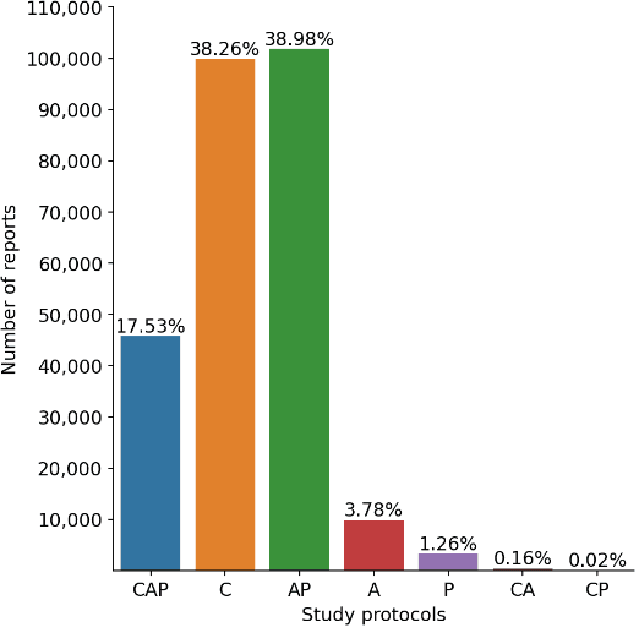
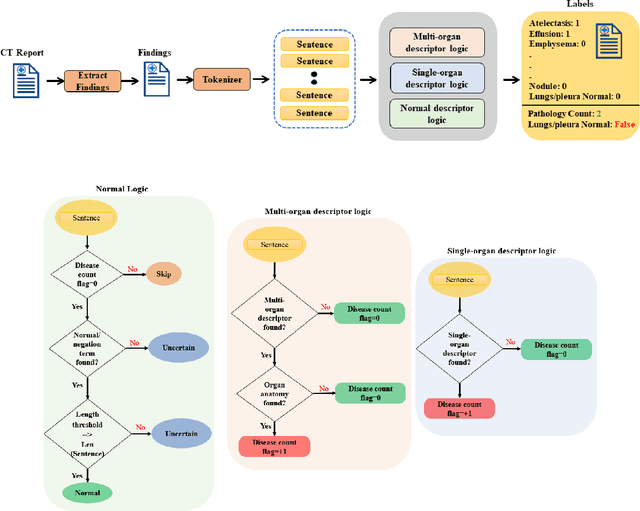
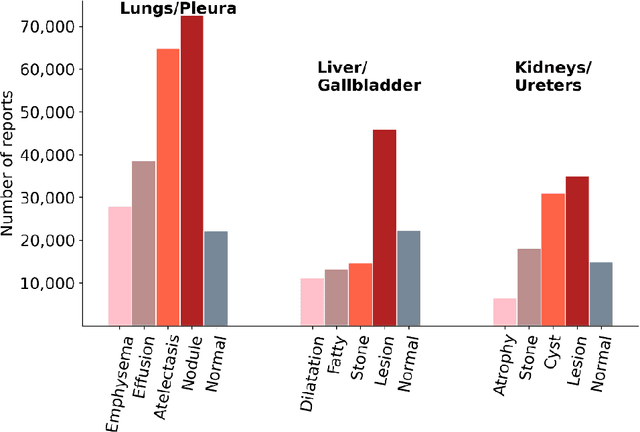
To develop a high throughput multi-label annotator for body Computed Tomography (CT) reports that can be applied to a variety of diseases, organs, and cases. First, we used a dictionary approach to develop a rule-based algorithm (RBA) for extraction of disease labels from radiology text reports. We targeted three organ systems (lungs/pleura, liver/gallbladder, kidneys/ureters) with four diseases per system based on their prevalence in our dataset. To expand the algorithm beyond pre-defined keywords, an attention-guided recurrent neural network (RNN) was trained using the RBA-extracted labels to classify the reports as being positive for one or more diseases or normal for each organ system. Confounding effects on model performance were evaluated using random or pre-trained embedding as well as different sizes of training datasets. Performance was evaluated using the receiver operating characteristic (ROC) area under the curve (AUC) against 2,158 manually obtained labels. Our model extracted disease labels from 261,229 radiology reports of 112,501 unique subjects. Pre-trained models outperformed random embedding across all diseases. As the training dataset size was reduced, performance was robust except for a few diseases with relatively small number of cases. Pre-trained Classification AUCs achieved > 0.95 for all five disease outcomes across all three organ systems. Our label-extracting pipeline was able to encompass a variety of cases and diseases by generalizing beyond strict rules with exceptional accuracy. As a framework, this model can be easily adapted to enable automated labeling of hospital-scale medical data sets for training image-based disease classifiers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge