Benjamin Danek
BioDSA-1K: Benchmarking Data Science Agents for Biomedical Research
May 22, 2025Abstract:Validating scientific hypotheses is a central challenge in biomedical research, and remains difficult for artificial intelligence (AI) agents due to the complexity of real-world data analysis and evidence interpretation. In this work, we present BioDSA-1K, a benchmark designed to evaluate AI agents on realistic, data-driven biomedical hypothesis validation tasks. BioDSA-1K consists of 1,029 hypothesis-centric tasks paired with 1,177 analysis plans, curated from over 300 published biomedical studies to reflect the structure and reasoning found in authentic research workflows. Each task includes a structured hypothesis derived from the original study's conclusions, expressed in the affirmative to reflect the language of scientific reporting, and one or more pieces of supporting evidence grounded in empirical data tables. While these hypotheses mirror published claims, they remain testable using standard statistical or machine learning methods. The benchmark enables evaluation along four axes: (1) hypothesis decision accuracy, (2) alignment between evidence and conclusion, (3) correctness of the reasoning process, and (4) executability of the AI-generated analysis code. Importantly, BioDSA-1K includes non-verifiable hypotheses: cases where the available data are insufficient to support or refute a claim, reflecting a common yet underexplored scenario in real-world science. We propose BioDSA-1K as a foundation for building and evaluating generalizable, trustworthy AI agents for biomedical discovery.
TrialPanorama: Database and Benchmark for Systematic Review and Design of Clinical Trials
May 22, 2025Abstract:Developing artificial intelligence (AI) for vertical domains requires a solid data foundation for both training and evaluation. In this work, we introduce TrialPanorama, a large-scale, structured database comprising 1,657,476 clinical trial records aggregated from 15 global sources. The database captures key aspects of trial design and execution, including trial setups, interventions, conditions, biomarkers, and outcomes, and links them to standard biomedical ontologies such as DrugBank and MedDRA. This structured and ontology-grounded design enables TrialPanorama to serve as a unified, extensible resource for a wide range of clinical trial tasks, including trial planning, design, and summarization. To demonstrate its utility, we derive a suite of benchmark tasks directly from the TrialPanorama database. The benchmark spans eight tasks across two categories: three for systematic review (study search, study screening, and evidence summarization) and five for trial design (arm design, eligibility criteria, endpoint selection, sample size estimation, and trial completion assessment). The experiments using five state-of-the-art large language models (LLMs) show that while general-purpose LLMs exhibit some zero-shot capability, their performance is still inadequate for high-stakes clinical trial workflows. We release TrialPanorama database and the benchmark to facilitate further research on AI for clinical trials.
InformGen: An AI Copilot for Accurate and Compliant Clinical Research Consent Document Generation
Apr 01, 2025Abstract:Leveraging large language models (LLMs) to generate high-stakes documents, such as informed consent forms (ICFs), remains a significant challenge due to the extreme need for regulatory compliance and factual accuracy. Here, we present InformGen, an LLM-driven copilot for accurate and compliant ICF drafting by optimized knowledge document parsing and content generation, with humans in the loop. We further construct a benchmark dataset comprising protocols and ICFs from 900 clinical trials. Experimental results demonstrate that InformGen achieves near 100% compliance with 18 core regulatory rules derived from FDA guidelines, outperforming a vanilla GPT-4o model by up to 30%. Additionally, a user study with five annotators shows that InformGen, when integrated with manual intervention, attains over 90% factual accuracy, significantly surpassing the vanilla GPT-4o model's 57%-82%. Crucially, InformGen ensures traceability by providing inline citations to source protocols, enabling easy verification and maintaining the highest standards of factual integrity.
A Perspective for Adapting Generalist AI to Specialized Medical AI Applications and Their Challenges
Oct 28, 2024
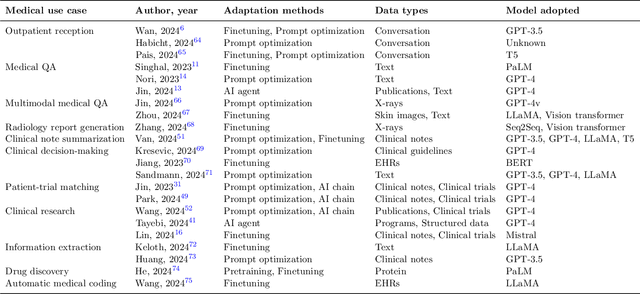

Abstract:The integration of Large Language Models (LLMs) into medical applications has sparked widespread interest across the healthcare industry, from drug discovery and development to clinical decision support, assisting telemedicine, medical devices, and healthcare insurance applications. This perspective paper aims to discuss the inner workings of building LLM-powered medical AI applications and introduces a comprehensive framework for their development. We review existing literature and outline the unique challenges of applying LLMs in specialized medical contexts. Additionally, we introduce a three-step framework to organize medical LLM research activities: 1) Modeling: breaking down complex medical workflows into manageable steps for developing medical-specific models; 2) Optimization: optimizing the model performance with crafted prompts and integrating external knowledge and tools, and 3) System engineering: decomposing complex tasks into subtasks and leveraging human expertise for building medical AI applications. Furthermore, we offer a detailed use case playbook that describes various LLM-powered medical AI applications, such as optimizing clinical trial design, enhancing clinical decision support, and advancing medical imaging analysis. Finally, we discuss various challenges and considerations for building medical AI applications with LLMs, such as handling hallucination issues, data ownership and compliance, privacy, intellectual property considerations, compute cost, sustainability issues, and responsible AI requirements.
Can Large Language Models Replace Data Scientists in Clinical Research?
Oct 28, 2024



Abstract:Data science plays a critical role in clinical research, but it requires professionals with expertise in coding and medical data analysis. Large language models (LLMs) have shown great potential in supporting medical tasks and performing well in general coding tests. However, these tests do not assess LLMs' ability to handle data science tasks in medicine, nor do they explore their practical utility in clinical research. To address this, we developed a dataset consisting of 293 real-world data science coding tasks, based on 39 published clinical studies, covering 128 tasks in Python and 165 tasks in R. This dataset simulates realistic clinical research scenarios using patient data. Our findings reveal that cutting-edge LLMs struggle to generate perfect solutions, frequently failing to follow input instructions, understand target data, and adhere to standard analysis practices. Consequently, LLMs are not yet ready to fully automate data science tasks. We benchmarked advanced adaptation methods and found two to be particularly effective: chain-of-thought prompting, which provides a step-by-step plan for data analysis, which led to a 60% improvement in code accuracy; and self-reflection, enabling LLMs to iteratively refine their code, yielding a 38% accuracy improvement. Building on these insights, we developed a platform that integrates LLMs into the data science workflow for medical professionals. In a user study with five medical doctors, we found that while LLMs cannot fully automate coding tasks, they significantly streamline the programming process. We found that 80% of their submitted code solutions were incorporated from LLM-generated code, with up to 96% reuse in some cases. Our analysis highlights the potential of LLMs, when integrated into expert workflows, to enhance data science efficiency in clinical research.
Accelerating Clinical Evidence Synthesis with Large Language Models
Jun 25, 2024Abstract:Automatic medical discovery by AI is a dream of many. One step toward that goal is to create an AI model to understand clinical studies and synthesize clinical evidence from the literature. Clinical evidence synthesis currently relies on systematic reviews of clinical trials and retrospective analyses from medical literature. However, the rapid expansion of publications presents challenges in efficiently identifying, summarizing, and updating evidence. We introduce TrialMind, a generative AI-based pipeline for conducting medical systematic reviews, encompassing study search, screening, and data extraction phases. We utilize large language models (LLMs) to drive each pipeline component while incorporating human expert oversight to minimize errors. To facilitate evaluation, we also create a benchmark dataset TrialReviewBench, a custom dataset with 870 annotated clinical studies from 25 meta-analysis papers across various medical treatments. Our results demonstrate that TrialMind significantly improves the literature review process, achieving high recall rates (0.897-1.000) in study searching from over 20 million PubMed studies and outperforming traditional language model embeddings-based methods in screening (Recall@20 of 0.227-0.246 vs. 0.000-0.102). Furthermore, our approach surpasses direct GPT-4 performance in result extraction, with accuracy ranging from 0.65 to 0.84. We also support clinical evidence synthesis in forest plots, as validated by eight human annotators who preferred TrialMind over the GPT-4 baseline with a winning rate of 62.5%-100% across the involved reviews. Our findings suggest that an LLM-based clinical evidence synthesis approach, such as TrialMind, can enable reliable and high-quality clinical evidence synthesis to improve clinical research efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge