Pengcheng Jiang
Rethinking the Reranker: Boundary-Aware Evidence Selection for Robust Retrieval-Augmented Generation
Feb 03, 2026Abstract:Retrieval-Augmented Generation (RAG) systems remain brittle under realistic retrieval noise, even when the required evidence appears in the top-K results. A key reason is that retrievers and rerankers optimize solely for relevance, often selecting either trivial, answer-revealing passages or evidence that lacks the critical information required to answer the question, without considering whether the evidence is suitable for the generator. We propose BAR-RAG, which reframes the reranker as a boundary-aware evidence selector that targets the generator's Goldilocks Zone -- evidence that is neither trivially easy nor fundamentally unanswerable for the generator, but is challenging yet sufficient for inference and thus provides the strongest learning signal. BAR-RAG trains the selector with reinforcement learning using generator feedback, and adopts a two-stage pipeline that fine-tunes the generator under the induced evidence distribution to mitigate the distribution mismatch between training and inference. Experiments on knowledge-intensive question answering benchmarks show that BAR-RAG consistently improves end-to-end performance under noisy retrieval, achieving an average gain of 10.3 percent over strong RAG and reranking baselines while substantially improving robustness. Code is publicly avaliable at https://github.com/GasolSun36/BAR-RAG.
Adaptation of Agentic AI
Dec 22, 2025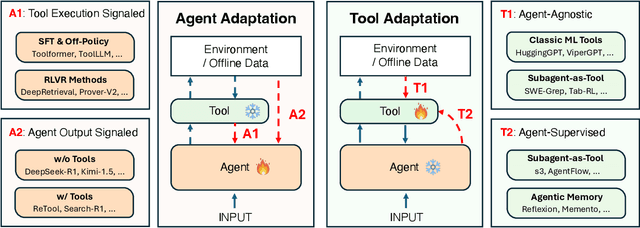
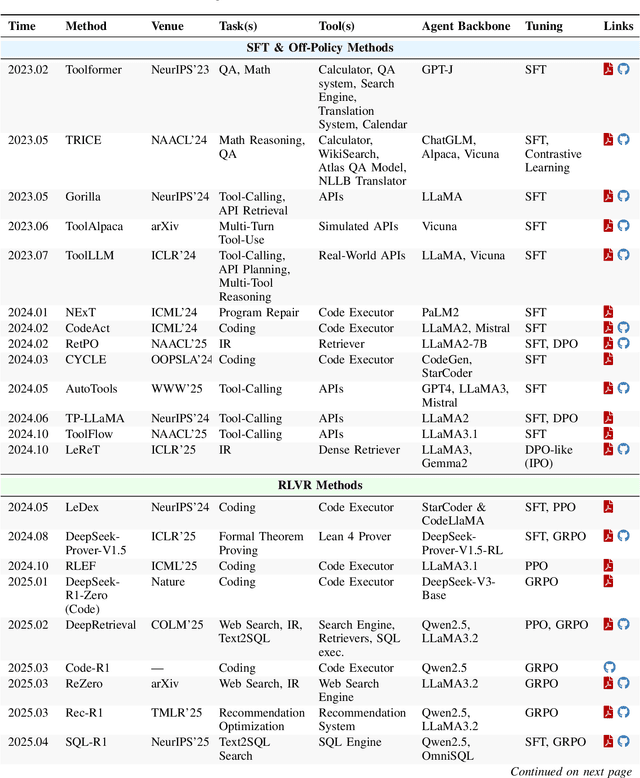
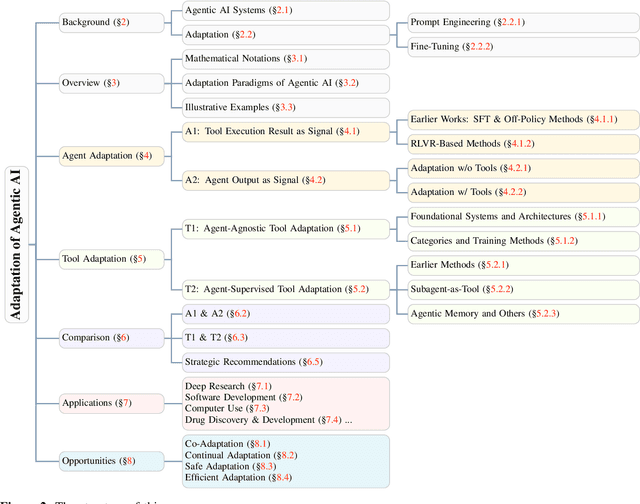
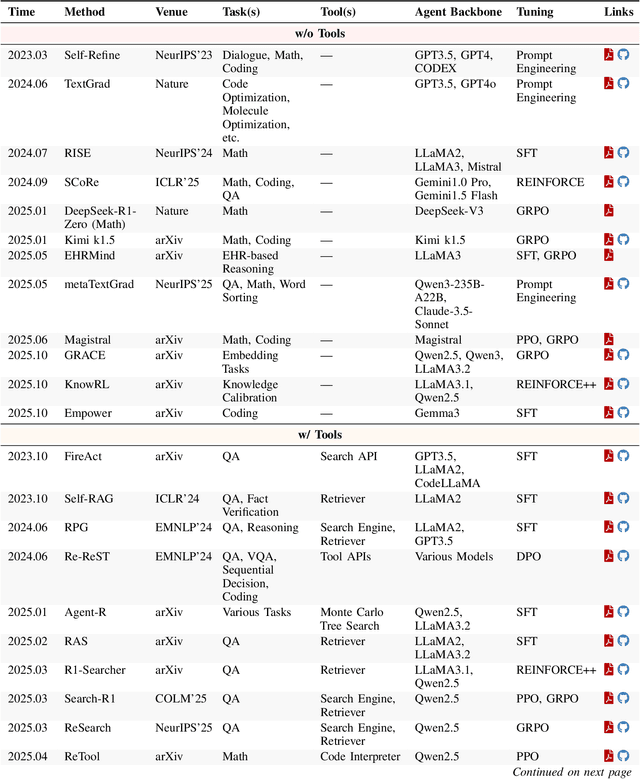
Abstract:Cutting-edge agentic AI systems are built on foundation models that can be adapted to plan, reason, and interact with external tools to perform increasingly complex and specialized tasks. As these systems grow in capability and scope, adaptation becomes a central mechanism for improving performance, reliability, and generalization. In this paper, we unify the rapidly expanding research landscape into a systematic framework that spans both agent adaptations and tool adaptations. We further decompose these into tool-execution-signaled and agent-output-signaled forms of agent adaptation, as well as agent-agnostic and agent-supervised forms of tool adaptation. We demonstrate that this framework helps clarify the design space of adaptation strategies in agentic AI, makes their trade-offs explicit, and provides practical guidance for selecting or switching among strategies during system design. We then review the representative approaches in each category, analyze their strengths and limitations, and highlight key open challenges and future opportunities. Overall, this paper aims to offer a conceptual foundation and practical roadmap for researchers and practitioners seeking to build more capable, efficient, and reliable agentic AI systems.
GRACE: Generative Representation Learning via Contrastive Policy Optimization
Oct 06, 2025Abstract:Prevailing methods for training Large Language Models (LLMs) as text encoders rely on contrastive losses that treat the model as a black box function, discarding its generative and reasoning capabilities in favor of static embeddings. We introduce GRACE (Generative Representation Learning via Contrastive Policy Optimization), a novel framework that reimagines contrastive signals not as losses to be minimized, but as rewards that guide a generative policy. In GRACE, the LLM acts as a policy that produces explicit, human-interpretable rationales--structured natural language explanations of its semantic understanding. These rationales are then encoded into high-quality embeddings via mean pooling. Using policy gradient optimization, we train the model with a multi-component reward function that maximizes similarity between query positive pairs and minimizes similarity with negatives. This transforms the LLM from an opaque encoder into an interpretable agent whose reasoning process is transparent and inspectable. On MTEB benchmark, GRACE yields broad cross category gains: averaged over four backbones, the supervised setting improves overall score by 11.5% over base models, and the unsupervised variant adds 6.9%, while preserving general capabilities. This work treats contrastive objectives as rewards over rationales, unifying representation learning with generation to produce stronger embeddings and transparent rationales. The model, data and code are available at https://github.com/GasolSun36/GRACE.
Topic Coverage-based Demonstration Retrieval for In-Context Learning
Sep 15, 2025Abstract:The effectiveness of in-context learning relies heavily on selecting demonstrations that provide all the necessary information for a given test input. To achieve this, it is crucial to identify and cover fine-grained knowledge requirements. However, prior methods often retrieve demonstrations based solely on embedding similarity or generation probability, resulting in irrelevant or redundant examples. In this paper, we propose TopicK, a topic coverage-based retrieval framework that selects demonstrations to comprehensively cover topic-level knowledge relevant to both the test input and the model. Specifically, TopicK estimates the topics required by the input and assesses the model's knowledge on those topics. TopicK then iteratively selects demonstrations that introduce previously uncovered required topics, in which the model exhibits low topical knowledge. We validate the effectiveness of TopicK through extensive experiments across various datasets and both open- and closed-source LLMs. Our source code is available at https://github.com/WonbinKweon/TopicK_EMNLP2025.
CC-RAG: Structured Multi-Hop Reasoning via Theme-Based Causal Graphs
Jun 11, 2025Abstract:Understanding cause and effect relationships remains a formidable challenge for Large Language Models (LLMs), particularly in specialized domains where reasoning requires more than surface-level correlations. Retrieval-Augmented Generation (RAG) improves factual accuracy, but standard RAG pipelines treat evidence as flat context, lacking the structure required to model true causal dependencies. We introduce Causal-Chain RAG (CC-RAG), a novel approach that integrates zero-shot triple extraction and theme-aware graph chaining into the RAG pipeline, enabling structured multi-hop inference. Given a domain specific corpus, CC-RAG constructs a Directed Acyclic Graph (DAG) of <cause, relation, effect> triples and uses forward/backward chaining to guide structured answer generation. Experiments on two real-world domains: Bitcoin price fluctuations and Gaucher disease, show that CC-RAG outperforms standard RAG and zero-shot LLMs in chain similarity, information density, and lexical diversity. Both LLM-as-a-Judge and human evaluations consistently favor CC-RAG. Our results demonstrate that explicitly modeling causal structure enables LLMs to generate more accurate and interpretable responses, especially in specialized domains where flat retrieval fails.
Zero-Shot Open-Schema Entity Structure Discovery
Jun 04, 2025Abstract:Entity structure extraction, which aims to extract entities and their associated attribute-value structures from text, is an essential task for text understanding and knowledge graph construction. Existing methods based on large language models (LLMs) typically rely heavily on predefined entity attribute schemas or annotated datasets, often leading to incomplete extraction results. To address these challenges, we introduce Zero-Shot Open-schema Entity Structure Discovery (ZOES), a novel approach to entity structure extraction that does not require any schema or annotated samples. ZOES operates via a principled mechanism of enrichment, refinement, and unification, based on the insight that an entity and its associated structure are mutually reinforcing. Experiments demonstrate that ZOES consistently enhances LLMs' ability to extract more complete entity structures across three different domains, showcasing both the effectiveness and generalizability of the method. These findings suggest that such an enrichment, refinement, and unification mechanism may serve as a principled approach to improving the quality of LLM-based entity structure discovery in various scenarios.
TrialPanorama: Database and Benchmark for Systematic Review and Design of Clinical Trials
May 22, 2025Abstract:Developing artificial intelligence (AI) for vertical domains requires a solid data foundation for both training and evaluation. In this work, we introduce TrialPanorama, a large-scale, structured database comprising 1,657,476 clinical trial records aggregated from 15 global sources. The database captures key aspects of trial design and execution, including trial setups, interventions, conditions, biomarkers, and outcomes, and links them to standard biomedical ontologies such as DrugBank and MedDRA. This structured and ontology-grounded design enables TrialPanorama to serve as a unified, extensible resource for a wide range of clinical trial tasks, including trial planning, design, and summarization. To demonstrate its utility, we derive a suite of benchmark tasks directly from the TrialPanorama database. The benchmark spans eight tasks across two categories: three for systematic review (study search, study screening, and evidence summarization) and five for trial design (arm design, eligibility criteria, endpoint selection, sample size estimation, and trial completion assessment). The experiments using five state-of-the-art large language models (LLMs) show that while general-purpose LLMs exhibit some zero-shot capability, their performance is still inadequate for high-stakes clinical trial workflows. We release TrialPanorama database and the benchmark to facilitate further research on AI for clinical trials.
s3: You Don't Need That Much Data to Train a Search Agent via RL
May 20, 2025Abstract:Retrieval-augmented generation (RAG) systems empower large language models (LLMs) to access external knowledge during inference. Recent advances have enabled LLMs to act as search agents via reinforcement learning (RL), improving information acquisition through multi-turn interactions with retrieval engines. However, existing approaches either optimize retrieval using search-only metrics (e.g., NDCG) that ignore downstream utility or fine-tune the entire LLM to jointly reason and retrieve-entangling retrieval with generation and limiting the real search utility and compatibility with frozen or proprietary models. In this work, we propose s3, a lightweight, model-agnostic framework that decouples the searcher from the generator and trains the searcher using a Gain Beyond RAG reward: the improvement in generation accuracy over naive RAG. s3 requires only 2.4k training samples to outperform baselines trained on over 70x more data, consistently delivering stronger downstream performance across six general QA and five medical QA benchmarks.
Taxonomy-guided Semantic Indexing for Academic Paper Search
Oct 25, 2024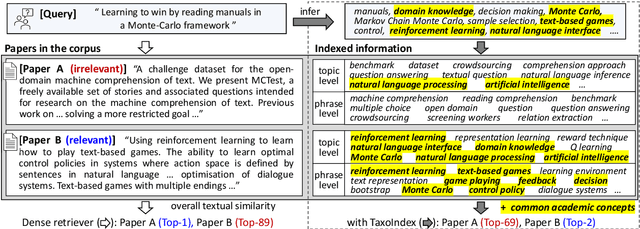
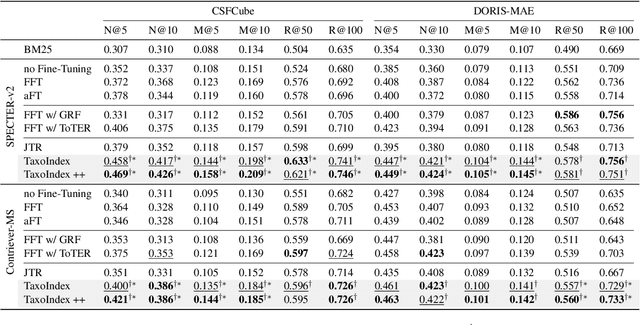
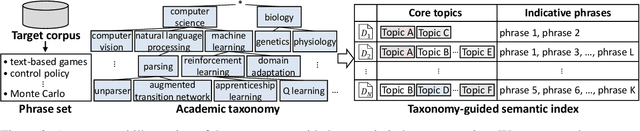
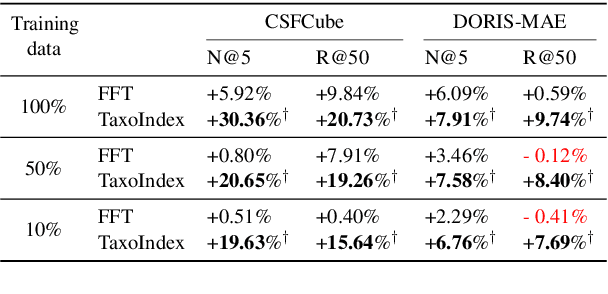
Abstract:Academic paper search is an essential task for efficient literature discovery and scientific advancement. While dense retrieval has advanced various ad-hoc searches, it often struggles to match the underlying academic concepts between queries and documents, which is critical for paper search. To enable effective academic concept matching for paper search, we propose Taxonomy-guided Semantic Indexing (TaxoIndex) framework. TaxoIndex extracts key concepts from papers and organizes them as a semantic index guided by an academic taxonomy, and then leverages this index as foundational knowledge to identify academic concepts and link queries and documents. As a plug-and-play framework, TaxoIndex can be flexibly employed to enhance existing dense retrievers. Extensive experiments show that TaxoIndex brings significant improvements, even with highly limited training data, and greatly enhances interpretability.
Reasoning-Enhanced Healthcare Predictions with Knowledge Graph Community Retrieval
Oct 06, 2024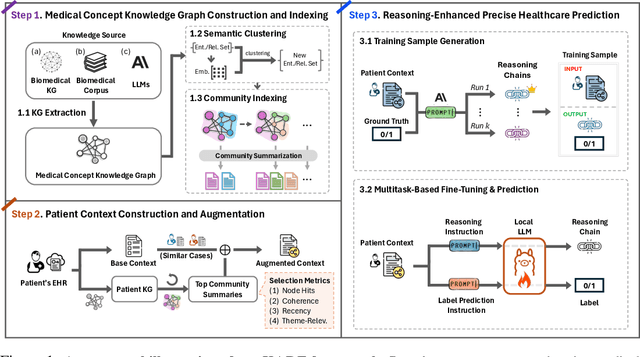
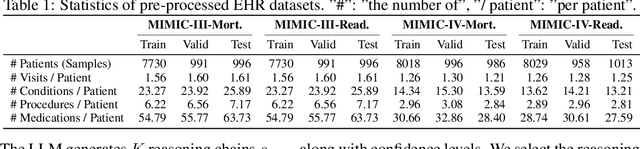
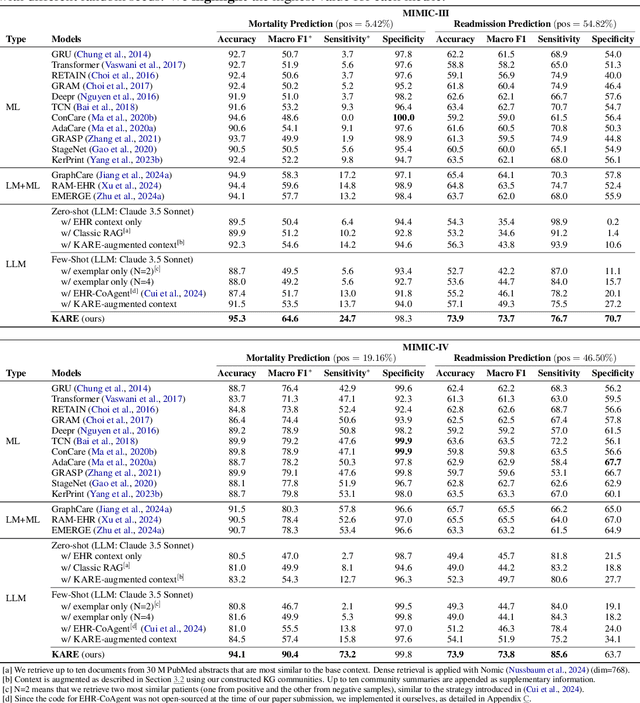
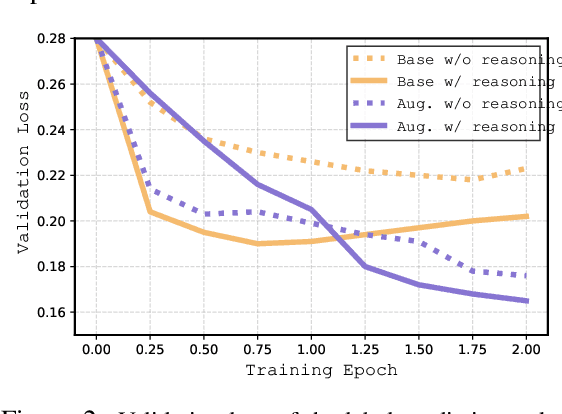
Abstract:Large language models (LLMs) have demonstrated significant potential in clinical decision support. Yet LLMs still suffer from hallucinations and lack fine-grained contextual medical knowledge, limiting their high-stake healthcare applications such as clinical diagnosis. Traditional retrieval-augmented generation (RAG) methods attempt to address these limitations but frequently retrieve sparse or irrelevant information, undermining prediction accuracy. We introduce KARE, a novel framework that integrates knowledge graph (KG) community-level retrieval with LLM reasoning to enhance healthcare predictions. KARE constructs a comprehensive multi-source KG by integrating biomedical databases, clinical literature, and LLM-generated insights, and organizes it using hierarchical graph community detection and summarization for precise and contextually relevant information retrieval. Our key innovations include: (1) a dense medical knowledge structuring approach enabling accurate retrieval of relevant information; (2) a dynamic knowledge retrieval mechanism that enriches patient contexts with focused, multi-faceted medical insights; and (3) a reasoning-enhanced prediction framework that leverages these enriched contexts to produce both accurate and interpretable clinical predictions. Extensive experiments demonstrate that KARE outperforms leading models by up to 10.8-15.0% on MIMIC-III and 12.6-12.7% on MIMIC-IV for mortality and readmission predictions. In addition to its impressive prediction accuracy, our framework leverages the reasoning capabilities of LLMs, enhancing the trustworthiness of clinical predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge