Jathurshan Pradeepkumar
ODEBrain: Continuous-Time EEG Graph for Modeling Dynamic Brain Networks
Feb 26, 2026Abstract:Modeling neural population dynamics is crucial for foundational neuroscientific research and various clinical applications. Conventional latent variable methods typically model continuous brain dynamics through discretizing time with recurrent architecture, which necessarily results in compounded cumulative prediction errors and failure of capturing instantaneous, nonlinear characteristics of EEGs. We propose ODEBRAIN, a Neural ODE latent dynamic forecasting framework to overcome these challenges by integrating spatio-temporal-frequency features into spectral graph nodes, followed by a Neural ODE modeling the continuous latent dynamics. Our design ensures that latent representations can capture stochastic variations of complex brain states at any given time point. Extensive experiments verify that ODEBRAIN can improve significantly over existing methods in forecasting EEG dynamics with enhanced robustness and generalization capabilities.
Making Conformal Predictors Robust in Healthcare Settings: a Case Study on EEG Classification
Feb 23, 2026Abstract:Quantifying uncertainty in clinical predictions is critical for high-stakes diagnosis tasks. Conformal prediction offers a principled approach by providing prediction sets with theoretical coverage guarantees. However, in practice, patient distribution shifts violate the i.i.d. assumptions underlying standard conformal methods, leading to poor coverage in healthcare settings. In this work, we evaluate several conformal prediction approaches on EEG seizure classification, a task with known distribution shift challenges and label uncertainty. We demonstrate that personalized calibration strategies can improve coverage by over 20 percentage points while maintaining comparable prediction set sizes. Our implementation is available via PyHealth, an open-source healthcare AI framework: https://github.com/sunlabuiuc/PyHealth.
Neural Signals Generate Clinical Notes in the Wild
Jan 29, 2026Abstract:Generating clinical reports that summarize abnormal patterns, diagnostic findings, and clinical interpretations from long-term EEG recordings remains labor-intensive. We curate a large-scale clinical EEG dataset with $9{,}922$ reports paired with approximately $11{,}000$ hours of EEG recordings from $9{,}048$ patients. We therefore develop CELM, the first clinical EEG-to-Language foundation model capable of summarizing long-duration, variable-length EEG recordings and performing end-to-end clinical report generation at multiple scales, including recording description, background activity, epileptiform abnormalities, events/seizures, and impressions. Experimental results show that, with patient history supervision, our method achieves $70\%$--$95\%$ average relative improvements in standard generation metrics (e.g., ROUGE-1 and METEOR) from $0.2$--$0.3$ to $0.4$--$0.6$. In the zero-shot setting without patient history, CELM attains generation scores in the range of $0.43$--$0.52$, compared to baselines of $0.17$--$0.26$. CELM integrates pretrained EEG foundation models with language models to enable scalable multimodal learning. We release our model and benchmark construction pipeline at [URL].
PyHealth 2.0: A Comprehensive Open-Source Toolkit for Accessible and Reproducible Clinical Deep Learning
Jan 23, 2026Abstract:Difficulty replicating baselines, high computational costs, and required domain expertise create persistent barriers to clinical AI research. To address these challenges, we introduce PyHealth 2.0, an enhanced clinical deep learning toolkit that enables predictive modeling in as few as 7 lines of code. PyHealth 2.0 offers three key contributions: (1) a comprehensive toolkit addressing reproducibility and compatibility challenges by unifying 15+ datasets, 20+ clinical tasks, 25+ models, 5+ interpretability methods, and uncertainty quantification including conformal prediction within a single framework that supports diverse clinical data modalities - signals, imaging, and electronic health records - with translation of 5+ medical coding standards; (2) accessibility-focused design accommodating multimodal data and diverse computational resources with up to 39x faster processing and 20x lower memory usage, enabling work from 16GB laptops to production systems; and (3) an active open-source community of 400+ members lowering domain expertise barriers through extensive documentation, reproducible research contributions, and collaborations with academic health systems and industry partners, including multi-language support via RHealth. PyHealth 2.0 establishes an open-source foundation and community advancing accessible, reproducible healthcare AI. Available at pip install pyhealth.
Prostate-VarBench: A Benchmark with Interpretable TabNet Framework for Prostate Cancer Variant Classification
Nov 12, 2025Abstract:Variants of Uncertain Significance (VUS) limit the clinical utility of prostate cancer genomics by delaying diagnosis and therapy when evidence for pathogenicity or benignity is incomplete. Progress is further limited by inconsistent annotations across sources and the absence of a prostate-specific benchmark for fair comparison. We introduce Prostate-VarBench, a curated pipeline for creating prostate-specific benchmarks that integrates COSMIC (somatic cancer mutations), ClinVar (expert-curated clinical variants), and TCGA-PRAD (prostate tumor genomics from The Cancer Genome Atlas) into a harmonized dataset of 193,278 variants supporting patient- or gene-aware splits to prevent data leakage. To ensure data integrity, we corrected a Variant Effect Predictor (VEP) issue that merged multiple transcript records, introducing ambiguity in clinical significance fields. We then standardized 56 interpretable features across eight clinically relevant tiers, including population frequency, variant type, and clinical context. AlphaMissense pathogenicity scores were incorporated to enhance missense variant classification and reduce VUS uncertainty. Building on this resource, we trained an interpretable TabNet model to classify variant pathogenicity, whose step-wise sparse masks provide per-case rationales consistent with molecular tumor board review practices. On the held-out test set, the model achieved 89.9% accuracy with balanced class metrics, and the VEP correction yields an 6.5% absolute reduction in VUS.
TrialPanorama: Database and Benchmark for Systematic Review and Design of Clinical Trials
May 22, 2025Abstract:Developing artificial intelligence (AI) for vertical domains requires a solid data foundation for both training and evaluation. In this work, we introduce TrialPanorama, a large-scale, structured database comprising 1,657,476 clinical trial records aggregated from 15 global sources. The database captures key aspects of trial design and execution, including trial setups, interventions, conditions, biomarkers, and outcomes, and links them to standard biomedical ontologies such as DrugBank and MedDRA. This structured and ontology-grounded design enables TrialPanorama to serve as a unified, extensible resource for a wide range of clinical trial tasks, including trial planning, design, and summarization. To demonstrate its utility, we derive a suite of benchmark tasks directly from the TrialPanorama database. The benchmark spans eight tasks across two categories: three for systematic review (study search, study screening, and evidence summarization) and five for trial design (arm design, eligibility criteria, endpoint selection, sample size estimation, and trial completion assessment). The experiments using five state-of-the-art large language models (LLMs) show that while general-purpose LLMs exhibit some zero-shot capability, their performance is still inadequate for high-stakes clinical trial workflows. We release TrialPanorama database and the benchmark to facilitate further research on AI for clinical trials.
Single-Channel EEG Tokenization Through Time-Frequency Modeling
Feb 22, 2025Abstract:We introduce TFM-Tokenizer, a novel tokenization framework tailored for EEG analysis that transforms continuous, noisy brain signals into a sequence of discrete, well-represented tokens for various EEG tasks. Conventional approaches typically rely on continuous embeddings and inter-channel dependencies, which are limited in capturing inherent EEG features such as temporally unpredictable patterns and diverse oscillatory waveforms. In contrast, we hypothesize that critical time-frequency features can be effectively captured from a single channel. By learning tokens that encapsulate these intrinsic patterns within a single channel, our approach yields a scalable tokenizer adaptable across diverse EEG settings. We integrate the TFM-Tokenizer with a transformer-based TFM-Encoder, leveraging established pretraining techniques from natural language processing, such as masked token prediction, followed by downstream fine-tuning for various EEG tasks. Experiments across four EEG datasets show that TFM-Token outperforms state-of-the-art methods. On TUEV, our approach improves balanced accuracy and Cohen's Kappa by 5% over baselines. Comprehensive analysis of the learned tokens demonstrates their ability to capture class-distinctive features, enhance frequency representation, and ability to encode time-frequency motifs into distinct tokens, improving interpretability.
Automatically Labeling $200B Life-Saving Datasets: A Large Clinical Trial Outcome Benchmark
Jun 13, 2024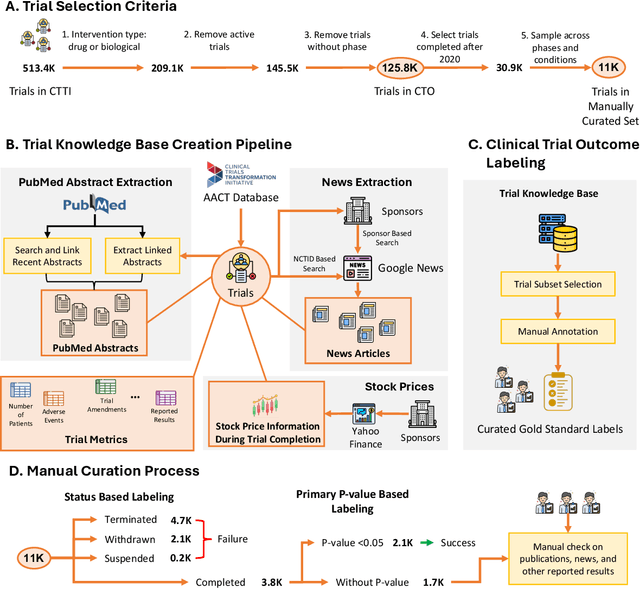
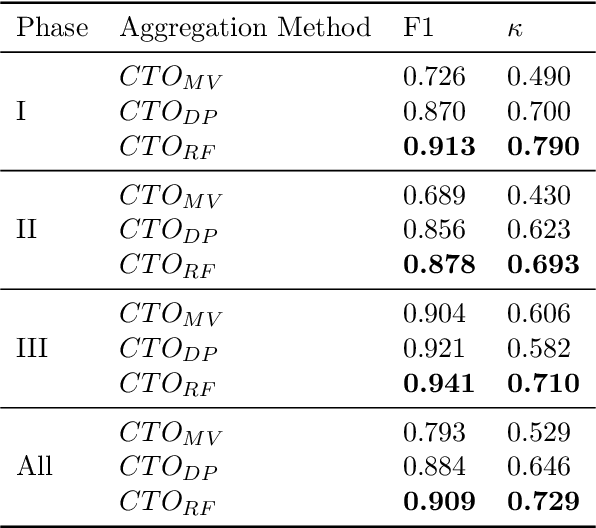
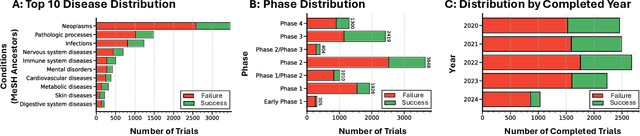
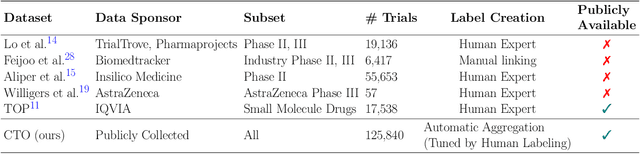
Abstract:The global cost of drug discovery and development exceeds $200 billion annually. The main results of drug discovery and development are the outcomes of clinical trials, which directly influence the regulatory approval of new drug candidates and ultimately affect patient outcomes. Despite their significance, large-scale, high-quality clinical trial outcome data are not readily available to the public. Suppose a large clinical trial outcome dataset is provided; machine learning researchers can potentially develop accurate prediction models using past trials and outcome labels, which could help prioritize and optimize therapeutic programs, ultimately benefiting patients. This paper introduces Clinical Trial Outcome (CTO) dataset, the largest trial outcome dataset with around 479K clinical trials, aggregating outcomes from multiple sources of weakly supervised labels, minimizing the noise from individual sources, and eliminating the need for human annotation. These sources include large language model (LLM) decisions on trial-related documents, news headline sentiments, stock prices of trial sponsors, trial linkages across phases, and other signals such as patient dropout rates and adverse events. CTO's labels show unprecedented agreement with supervised clinical trial outcome labels from test split of the supervised TOP dataset, with a 91 F1.
Contrastive Deep Encoding Enables Uncertainty-aware Machine-learning-assisted Histopathology
Sep 13, 2023



Abstract:Deep neural network models can learn clinically relevant features from millions of histopathology images. However generating high-quality annotations to train such models for each hospital, each cancer type, and each diagnostic task is prohibitively laborious. On the other hand, terabytes of training data -- while lacking reliable annotations -- are readily available in the public domain in some cases. In this work, we explore how these large datasets can be consciously utilized to pre-train deep networks to encode informative representations. We then fine-tune our pre-trained models on a fraction of annotated training data to perform specific downstream tasks. We show that our approach can reach the state-of-the-art (SOTA) for patch-level classification with only 1-10% randomly selected annotations compared to other SOTA approaches. Moreover, we propose an uncertainty-aware loss function, to quantify the model confidence during inference. Quantified uncertainty helps experts select the best instances to label for further training. Our uncertainty-aware labeling reaches the SOTA with significantly fewer annotations compared to random labeling. Last, we demonstrate how our pre-trained encoders can surpass current SOTA for whole-slide image classification with weak supervision. Our work lays the foundation for data and task-agnostic pre-trained deep networks with quantified uncertainty.
A Knowledge Distillation Framework For Enhancing Ear-EEG Based Sleep Staging With Scalp-EEG Data
Oct 27, 2022Abstract:Sleep plays a crucial role in the well-being of human lives. Traditional sleep studies using Polysomnography are associated with discomfort and often lower sleep quality caused by the acquisition setup. Previous works have focused on developing less obtrusive methods to conduct high-quality sleep studies, and ear-EEG is among popular alternatives. However, the performance of sleep staging based on ear-EEG is still inferior to scalp-EEG based sleep staging. In order to address the performance gap between scalp-EEG and ear-EEG based sleep staging, we propose a cross-modal knowledge distillation strategy, which is a domain adaptation approach. Our experiments and analysis validate the effectiveness of the proposed approach with existing architectures, where it enhances the accuracy of the ear-EEG based sleep staging by 3.46% and Cohen's kappa coefficient by a margin of 0.038.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge