Xihao Piao
Single-Channel EEG Tokenization Through Time-Frequency Modeling
Feb 22, 2025Abstract:We introduce TFM-Tokenizer, a novel tokenization framework tailored for EEG analysis that transforms continuous, noisy brain signals into a sequence of discrete, well-represented tokens for various EEG tasks. Conventional approaches typically rely on continuous embeddings and inter-channel dependencies, which are limited in capturing inherent EEG features such as temporally unpredictable patterns and diverse oscillatory waveforms. In contrast, we hypothesize that critical time-frequency features can be effectively captured from a single channel. By learning tokens that encapsulate these intrinsic patterns within a single channel, our approach yields a scalable tokenizer adaptable across diverse EEG settings. We integrate the TFM-Tokenizer with a transformer-based TFM-Encoder, leveraging established pretraining techniques from natural language processing, such as masked token prediction, followed by downstream fine-tuning for various EEG tasks. Experiments across four EEG datasets show that TFM-Token outperforms state-of-the-art methods. On TUEV, our approach improves balanced accuracy and Cohen's Kappa by 5% over baselines. Comprehensive analysis of the learned tokens demonstrates their ability to capture class-distinctive features, enhance frequency representation, and ability to encode time-frequency motifs into distinct tokens, improving interpretability.
A Unified Energy Management Framework for Multi-Timescale Forecasting in Smart Grids
Nov 22, 2024



Abstract:Accurate forecasting of the electrical load, such as the magnitude and the timing of peak power, is crucial to successful power system management and implementation of smart grid strategies like demand response and peak shaving. In multi-time-scale optimization scheduling, rolling optimization is a common solution. However, rolling optimization needs to consider the coupling of different optimization objectives across time scales. It is challenging to accurately capture the mid- and long-term dependencies in time series data. This paper proposes Multi-pofo, a multi-scale power load forecasting framework, that captures such dependency via a novel architecture equipped with a temporal positional encoding layer. To validate the effectiveness of the proposed model, we conduct experiments on real-world electricity load data. The experimental results show that our approach outperforms compared to several strong baseline methods.
FredNormer: Frequency Domain Normalization for Non-stationary Time Series Forecasting
Oct 06, 2024Abstract:Recent normalization-based methods have shown great success in tackling the distribution shift issue, facilitating non-stationary time series forecasting. Since these methods operate in the time domain, they may fail to fully capture the dynamic patterns that are more apparent in the frequency domain, leading to suboptimal results. This paper first theoretically analyzes how normalization methods affect frequency components. We prove that the current normalization methods that operate in the time domain uniformly scale non-zero frequencies, and thus, they struggle to determine components that contribute to more robust forecasting. Therefore, we propose FredNormer, which observes datasets from a frequency perspective and adaptively up-weights the key frequency components. To this end, FredNormer consists of two components: a statistical metric that normalizes the input samples based on their frequency stability and a learnable weighting layer that adjusts stability and introduces sample-specific variations. Notably, FredNormer is a plug-and-play module, which does not compromise the efficiency compared to existing normalization methods. Extensive experiments show that FredNormer improves the averaged MSE of backbone forecasting models by 33.3% and 55.3% on the ETTm2 dataset. Compared to the baseline normalization methods, FredNormer achieves 18 top-1 results and 6 top-2 results out of 28 settings.
CMOB: Large-Scale Cancer Multi-Omics Benchmark with Open Datasets, Tasks, and Baselines
Sep 02, 2024Abstract:Machine learning has shown great potential in the field of cancer multi-omics studies, offering incredible opportunities for advancing precision medicine. However, the challenges associated with dataset curation and task formulation pose significant hurdles, especially for researchers lacking a biomedical background. Here, we introduce the CMOB, the first large-scale cancer multi-omics benchmark integrates the TCGA platform, making data resources accessible and usable for machine learning researchers without significant preparation and expertise.To date, CMOB includes a collection of 20 cancer multi-omics datasets covering 32 cancers, accompanied by a systematic data processing pipeline. CMOB provides well-processed dataset versions to support 20 meaningful tasks in four studies, with a collection of benchmarks. We also integrate CMOB with two complementary resources and various biological tools to explore broader research avenues.All resources are open-accessible with user-friendly and compatible integration scripts that enable non-experts to easily incorporate this complementary information for various tasks. We conduct extensive experiments on selected datasets to offer recommendations on suitable machine learning baselines for specific applications. Through CMOB, we aim to facilitate algorithmic advances and hasten the development, validation, and clinical translation of machine-learning models for personalized cancer treatments. CMOB is available on GitHub (\url{https://github.com/chenzRG/Cancer-Multi-Omics-Benchmark}).
$\mathbb{BEHR}$NOULLI: A Binary EHR Data-Oriented Medication Recommendation System
Aug 18, 2024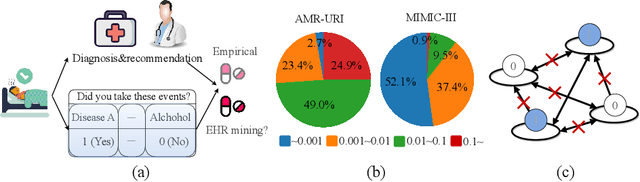
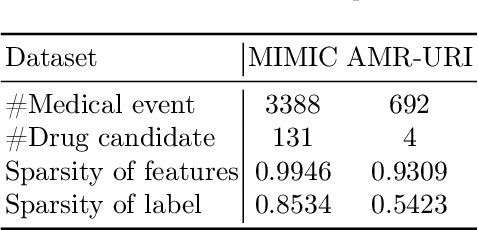
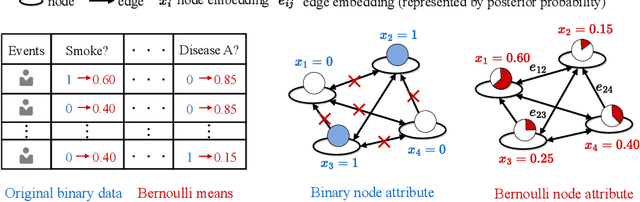

Abstract:The medical community believes binary medical event outcomes in EHR data contain sufficient information for making a sensible recommendation. However, there are two challenges to effectively utilizing such data: (1) modeling the relationship between massive 0,1 event outcomes is difficult, even with expert knowledge; (2) in practice, learning can be stalled by the binary values since the equally important 0 entries propagate no learning signals. Currently, there is a large gap between the assumed sufficient information and the reality that no promising results have been shown by utilizing solely the binary data: visiting or secondary information is often necessary to reach acceptable performance. In this paper, we attempt to build the first successful binary EHR data-oriented drug recommendation system by tackling the two difficulties, making sensible drug recommendations solely using the binary EHR medical records. To this end, we take a statistical perspective to view the EHR data as a sample from its cohorts and transform them into continuous Bernoulli probabilities. The transformed entries not only model a deterministic binary event with a distribution but also allow reflecting \emph{event-event} relationship by conditional probability. A graph neural network is learned on top of the transformation. It captures event-event correlations while emphasizing \emph{event-to-patient} features. Extensive results demonstrate that the proposed method achieves state-of-the-art performance on large-scale databases, outperforming baseline methods that use secondary information by a large margin. The source code is available at \url{https://github.com/chenzRG/BEHRMecom}
Fredformer: Frequency Debiased Transformer for Time Series Forecasting
Jun 14, 2024

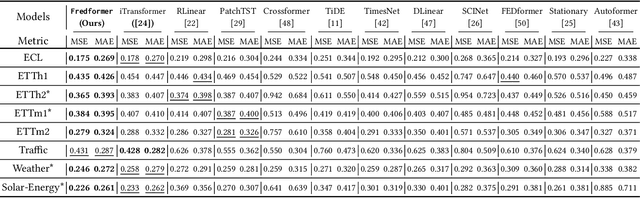

Abstract:The Transformer model has shown leading performance in time series forecasting. Nevertheless, in some complex scenarios, it tends to learn low-frequency features in the data and overlook high-frequency features, showing a frequency bias. This bias prevents the model from accurately capturing important high-frequency data features. In this paper, we undertook empirical analyses to understand this bias and discovered that frequency bias results from the model disproportionately focusing on frequency features with higher energy. Based on our analysis, we formulate this bias and propose Fredformer, a Transformer-based framework designed to mitigate frequency bias by learning features equally across different frequency bands. This approach prevents the model from overlooking lower amplitude features important for accurate forecasting. Extensive experiments show the effectiveness of our proposed approach, which can outperform other baselines in different real-world time-series datasets. Furthermore, we introduce a lightweight variant of the Fredformer with an attention matrix approximation, which achieves comparable performance but with much fewer parameters and lower computation costs. The code is available at: https://github.com/chenzRG/Fredformer
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge